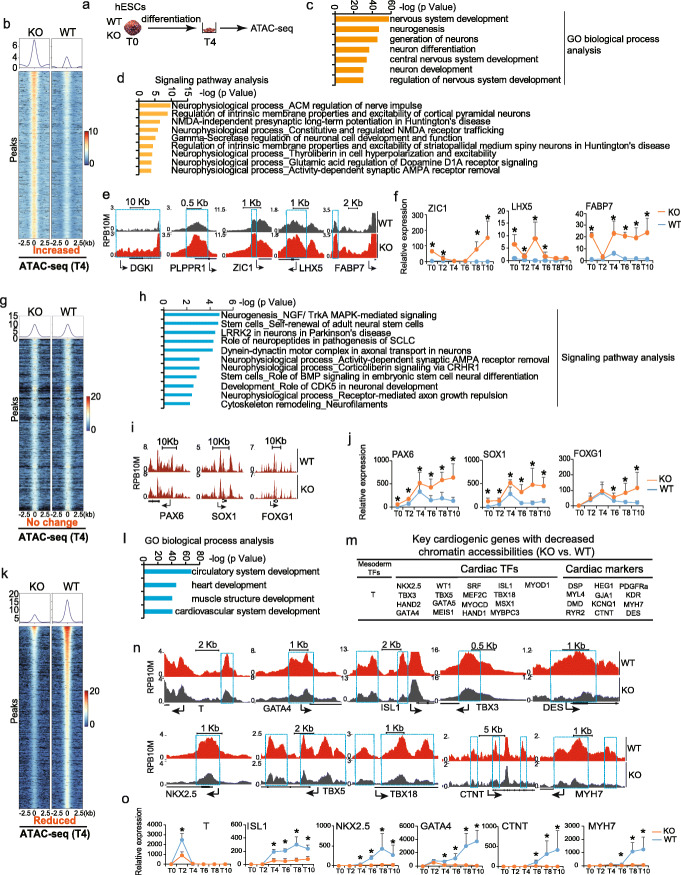
Fig. 5.
Loss-of-ARID1A leads to changes of chromatin accessibility. a Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) was performed after 4 days differentiation of WT and ARID1A−/− hESCs. b Heatmap and pileup of ATAC-seq signal for genes with increased chromatin accessibility (KO vs. WT). c, d GO biological process (c) and signaling pathway enrichment (d) analysis of genes with increased chromatin accessibility (KO vs. WT). The analysis of GO biological process and signaling pathways was performed by Metacore software. e Genome views of neurogenic genes with increased chromatin accessibility (KO vs. WT) close to transcription start site (TSS). Arrows showed the transcriptional direction. f qRT-PCR detection of neurogenic gene expression profiles during cardiac differentiation of hESCs. g Heatmap and pileup of ATAC-seq signals for genes without significant changes on chromatin accessibility (KO vs. WT). h Signaling pathways enriched in genes with no-changed chromatin accessibility (KO vs. WT). The analysis of signaling pathways was performed by Metacore software. i Genome views of neurogenic genes without significant changes on chromatin accessibility (KO vs. WT). j qRT-PCR detection of neurogenic gene expression profiles during cardiac differentiation of hESCs. k Heatmap and pileup of ATAC-seq signals for genes with reduced chromatin accessibilities (KO vs. WT). l GO biological process analysis of genes with reduced chromatin accessibilities (KO vs. WT). The analysis of GO biological process was performed by Metacore software. m List of essential cardiogenic and cardiac functional genes showing decreased chromatin accessibility in KO cells (KO vs. WT). n Genome views of some important cardiogenic genes with decreased chromatin accessibility in KO cells compared to WT. Arrows showed the transcriptional direction. o qRT-PCR detection of cardiogenic gene expression profiles during cardiac differentiation of hESCs. All bars in RT-qPCR data are shown as mean ± SD. n = 3, *p < 0.05 (KO vs. WT, an unpaired two-tailed t test with Welch’s correction)