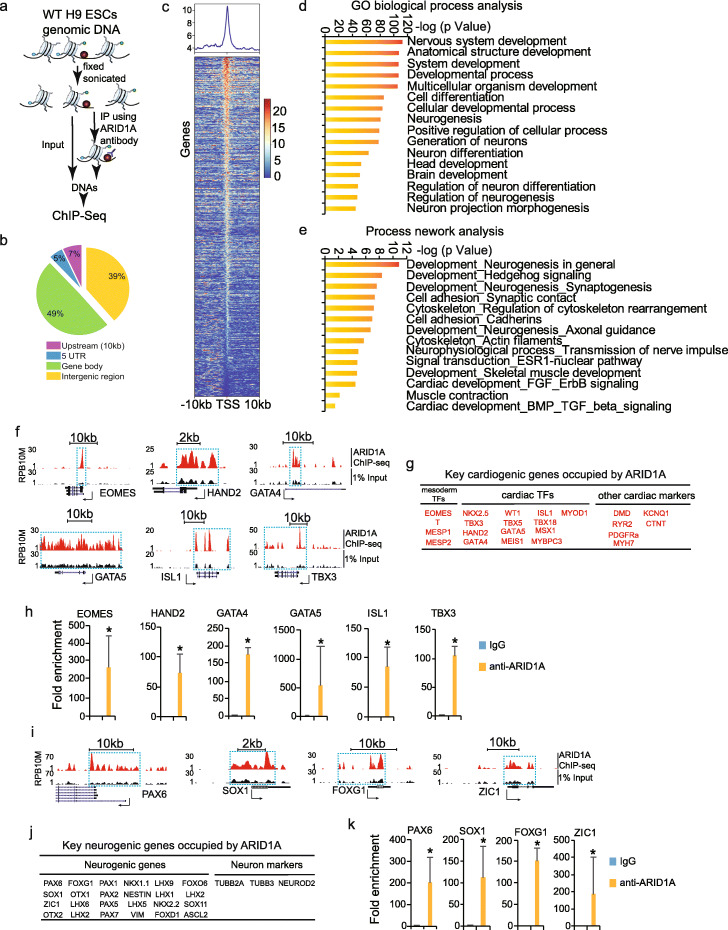
Fig. 6.
ChIP-seq reveals genome-wide ARID1A occupancies on promoters of cardiogenic and neurogenic genes. a Chromatin immunoprecipitation (ChIP) assays with sequencing (ChIP-Seq) was performed in WT H9 hESCs. b Distribution of ARID1A ChIP-seq-enriched peaks across human genome. c Relative enrichment levels of ARID1A on transcription start site (TSS) of coding genes in hESCs. Heatmaps were ranked by ARID1A enrichment. d GO biological process analysis of genes enriched by ARID1A ChIP-seq. The analysis of GO biological process was performed by Metacore software. e Process network analysis of genes enriched by ARID1A ChIP-seq. The analysis of process networks was performed by Metacore software. f Example genome views of ARID1A occupancies on mesodermal TF EOMES, cardiac TFs HAND2, GATA4, GATA5, ISL1, and TBX3. g List of essential cardiogenic and cardiac functional genes occupied by ARID1A. h ChIP-qPCR validation of cardiogenic gene promoters occupied by ARID1A. i Example genome views of ARID1A occupancies on neurogenic genes. j List of essential neurogenic and neural marker genes occupied by ARID1A. k ChIP-qPCR validation of neurogenic gene promoters occupied by ARID1A. All bars in ChIP-QPCR data are shown as mean ± SD. n = 3, *p < 0.05 (anti-ARID1A vs. IgG, an unpaired two-tailed t test)