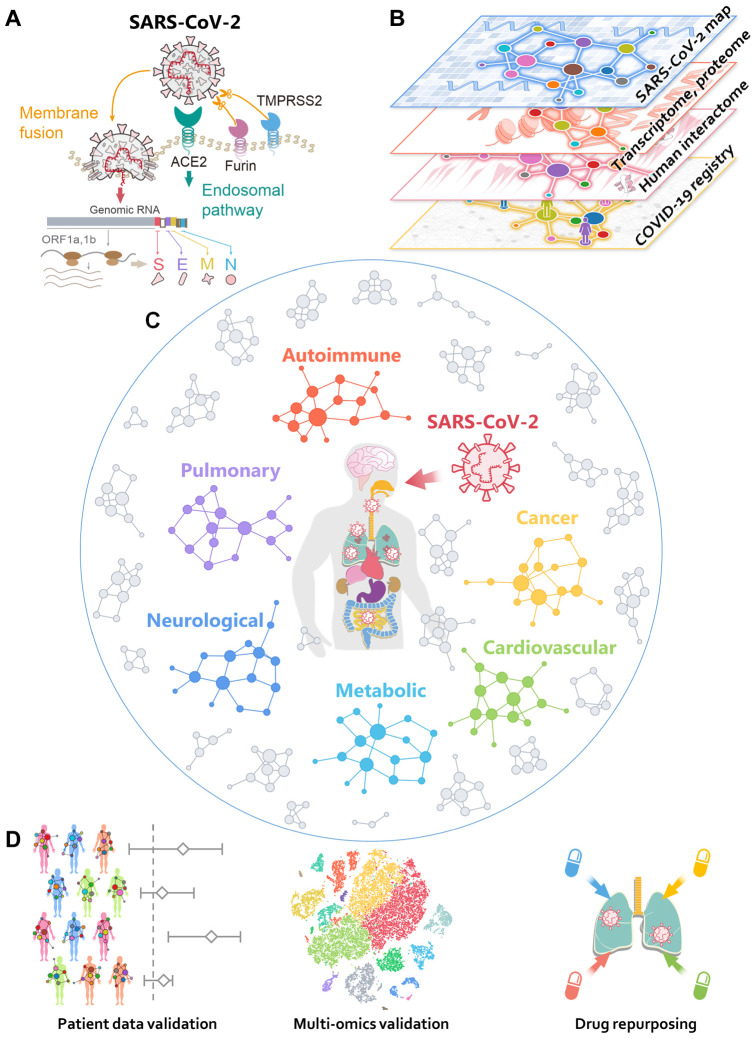
Fig. 1. Overall workflow of this study.
(A) A diagram illustrating the basic pathogenesis of SARS-CoV-2. (B) A diagram illustrating how to build a global interactome map for SARS-CoV-2. We compiled the SARS-CoV-2 human target gene/protein sets from multi-omics data from transcriptome, proteome, and human interactome, and validated network-based findings using patient data from the COVID-19 registry. (C) A diagram illustrating network-based measure of disease manifestations associated with COVID-19. We systematically evaluated the network proximities of the SARS-CoV-2 human target genes/proteins with 64 diseases across six main categories: autoimmune, cancer, cardiovascular, metabolic, neurological, and pulmonary. (D) A workflow illustrating validation of network-based findings. We performed single-cell analyses to further investigate the underlying mechanisms of COVID-19 with asthma and inflammatory bowel disease. We prioritized nearly 3,000 FDA approved/investigational drugs for their potential anti-SARS-CoV-2 effects from network-based findings and validated drug-COVID-19 outcomes using the institutional review board-approved COVID-19 patient registry.