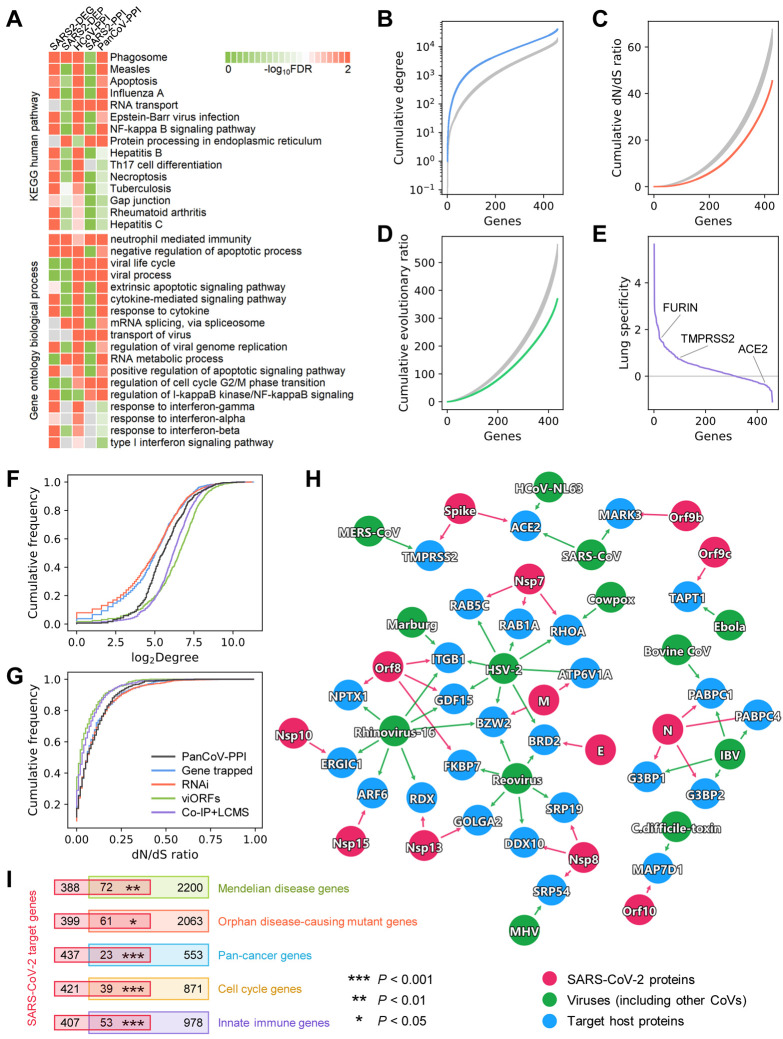
Fig. 2. Network and biological characteristics of the SARS-CoV-2 interactome map.
(A) Pathway and gene ontology (biological process) enrichment analysis results of the SARS-CoV-2 host genes/proteins across five different data sets. We assembled five gene/protein data sets from SARS-CoV-2 host protein-protein interactions, transcriptomics, and proteomics (Table S1). (B, C, D) Network and biological characteristics of the SARS-CoV-2 host genes/proteins. The proteins in PanCoV-PPI have higher node degrees (B), lower non-synonymous to synonymous substitutions (dN/dS) ratios (C), and lower evolutionary ratios (D) compared to randomly selected proteins (grey, mean ± standard deviation of 100 repeats). (E) Among the 460 proteins in PanCoV-PPI, 450 (98%) are expressed in lungs, and 317 (69%) are lung specific expression (Z > 0). (F, G) The distribution of the node degrees in the human interactome and dN/dS ratios of PanCoV-PPI and four published virus-related host protein sets. (H) The shared target human proteins (blue) of SARS-CoV-2 (red) and other viruses (green). The detailed data are provided in Table S2. (I) SARS-CoV-2 target proteins overlap significantly with disease-associated genes (Mendelian disease and Orphan disease), cancer genes, cell cycle genes, and innate immune genes.