Figure 1.
Expression of the Minimal DENV Replicase Does Not Suffice to Induce VP Formation
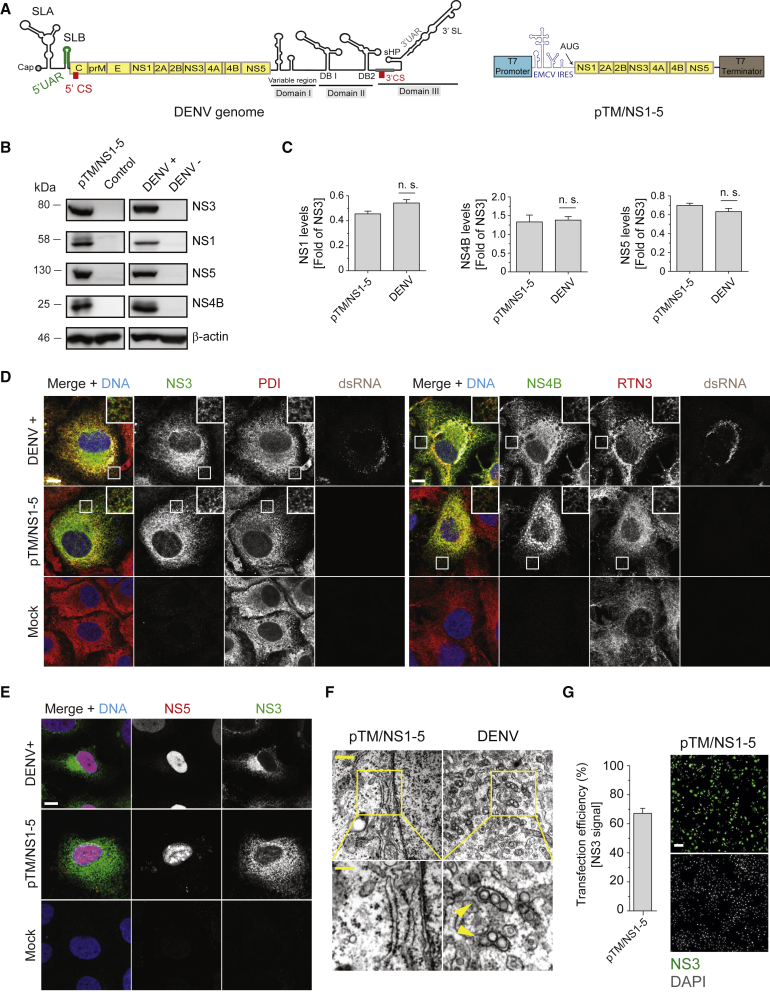
(A) Schematic representation of the DENV genome organization (left) and the T7 RNA polymerase-driven expression construct encoding the minimal DENV replicase NS1-5 (pTM/NS1-5; right panel). SL, stem-loop; UAR, upstream of AUG region; CS, cyclization sequence; DB, dumbbell; sHP, short-harpin.
(B) Huh7/Lunet-T7 cells were either transfected with pTM/NS1-5 for 20 h or infected with DENV (MOI = 5) for 48 h before being lysed and subjected to western blot analysis (left and right panels, respectively). β-actin was used as loading control.
(C) Relative abundance of viral proteins was determined by densitometry of the western blots, and values obtained for NS1, NS4B, or NS5 were normalized to NS3 expression levels. Values represent mean and standard error of three independent experiments. n.s., not significant.
(D) Cells were infected with DENV (upper row), transfected with the pTM/NS1-5 construct (middle row), or left untreated (bottom row) and fixed for immunofluorescence analysis after 48 h (infection) or 20 h (transfection and mock). PDI (protein disulfide isomerase) and RTN3 (reticulon 3) signals serve as ER makers. Scale bars: 10 μm.
(E) Nuclear localization of NS5 in DENV-infected or pTM/NS1-5-transfected cells. Scale bar: 10 μm.
(F) DENV-infected or pTM/NS1-5-transfected cells were fixed after 48 h or 20 h, respectively, and processed for TEM analysis. Lower panels are magnifications of areas highlighted with yellow squares in the upper panels. Yellow arrowheads show individual VPs. Scale bars: 500 and 200 nm in upper and lower panels, respectively.
(G) Transfection efficiency as determined by immunofluorescence staining of NS3. Error bars (left graph) represent the SEM of three independent experiments. Scale bar: 100 μm.