Figure 5.
Importance of the 3′ SL for DENV RO Formation
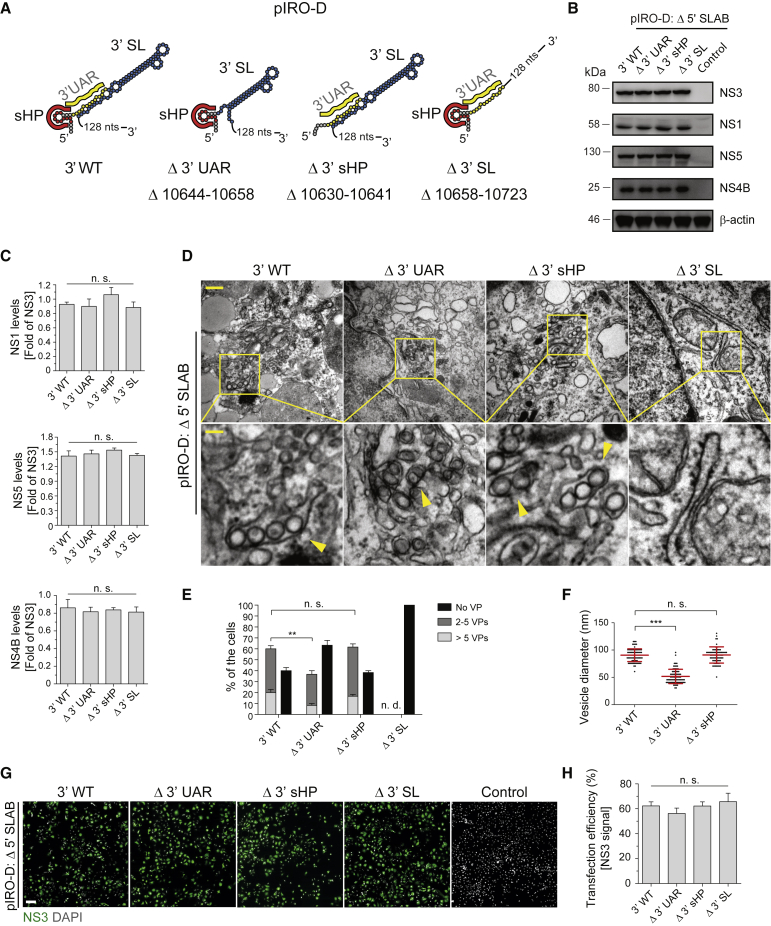
(A) Schematic representation of 3′ UTR truncations introduced into the pIRO-D Δ 5′ SLAB construct. Numbers refer to the deleted nucleotide positions of the DENV 2 genome (GenBank accession number NC_001474.2).
(B) Analysis of viral protein production in Huh7/Lunet-T7 cells 20 h after transfection with given constructs. β-actin served as sample loading control.
(C) Relative abundance of viral proteins was determined by densitometry, and levels of NS1, NS4B, and NS5 were normalized to NS3 signal. Values represent mean and standard error of three independent experiments. n.s., not significant.
(D) TEM images of Huh7/Lunet-T7 cells transfected with indicated 3′ UTR mutants or the 3′ WT reference construct. Cells were transfected and, after 20 h, fixed, processed, and embedded in resin for sectioning. Upper panel scale bar: 500 nm. Lower panels are magnifications of yellow squared areas in the upper panel images. Yellow arrowheads indicate individual VPs. Lower panel scale bar: 100 nm.
(E) For each condition, VPs contained in whole-cell sections from 20 cells or, in the case of the Δ 3′ SL mutant, 40 cells were counted. Means ± SEM from three independent experiments are shown. n.s., not significant; ∗∗p < 0.01.
(F) Vesicle diameters were measured by analyzing 25 vesicles per experimental condition using Fiji software. Means ± SEM from three independent experiments are given. n.s., not significant; ∗∗∗p < 0.001.
(G) Detection of NS3 by immunofluorescence microscopy to determine transfection efficiency for the indicated constructs. Nuclear DNA was stained with DAPI. Scale bar: 100 μm.
(H) Transfection efficiencies were quantified by counting NS3-containing cells and normalization to the total number of cells. Error bars represent the standard error of three independent experiments. n.s., not significant.
Note that percentages given in (E) refer to all cells analyzed by TEM, but only ~60% of them had been transfected (H); therefore, not more than 60% of cells can contain VPs.