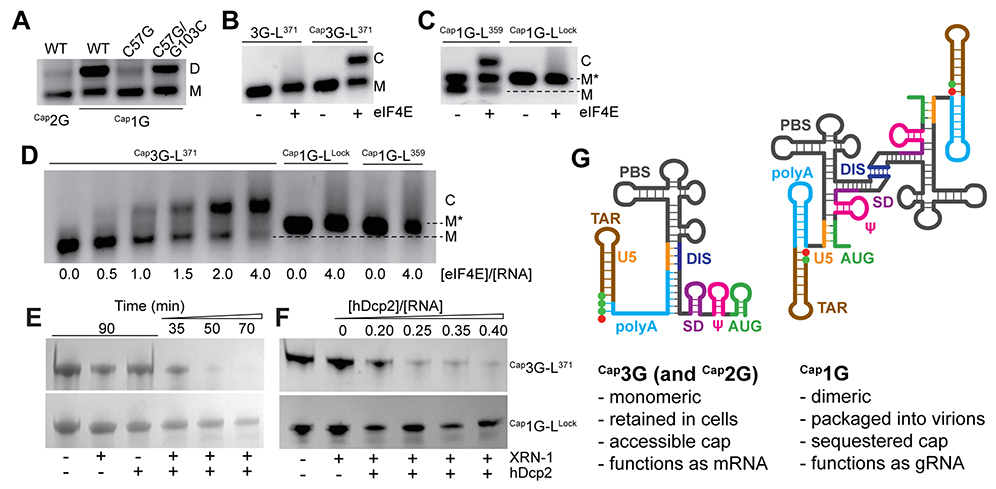
Fig. 4.
Influence of 5’-guanosine number on RNA function. (A) Disruption of a single base pair (C57-G103) by C57 to G mutagenesis disrupts Cap1G-L371 dimerization. Compensatory G103C substitution substantially restores dimerization. (B) eIF4E binds Cap3G-L371 (C denotes the eIF4E:RNA complex) but not the non-capped RNA. (C) At low ionic strength, eIF4E binds the M conformer of Cap1G-L359, butnotM* or the Cap1G-LLock construct. (D) Similar results were obtained in PI buffer. (E-F) The 5’-RNA exonuclease (XRN-1) and decapping enzyme (hDcp2) are independently unable to degrade Cap1G- or Cap3G-leader RNAs. In the presence of both enzymes, The Cap1G-LLock resists degradation over time (E) and with increasing hDcp2 (F) compared to the cap-exposed Cap3G-L371 leader. (G) Mechanism for transcriptional control of HIV-1 RNA function. Capped RNAs containing two or three 5’-guanosines adopt a monomeric structure that exposes the cap and enables RNA processing and metabolism, whereas those with a single capped G adopt a cap-sequestered conformation that promotes dimerization and packaging.