Figure 3. AcrIIA1 Inhibits Cas9 to Protect Prophages During Lysogeny.
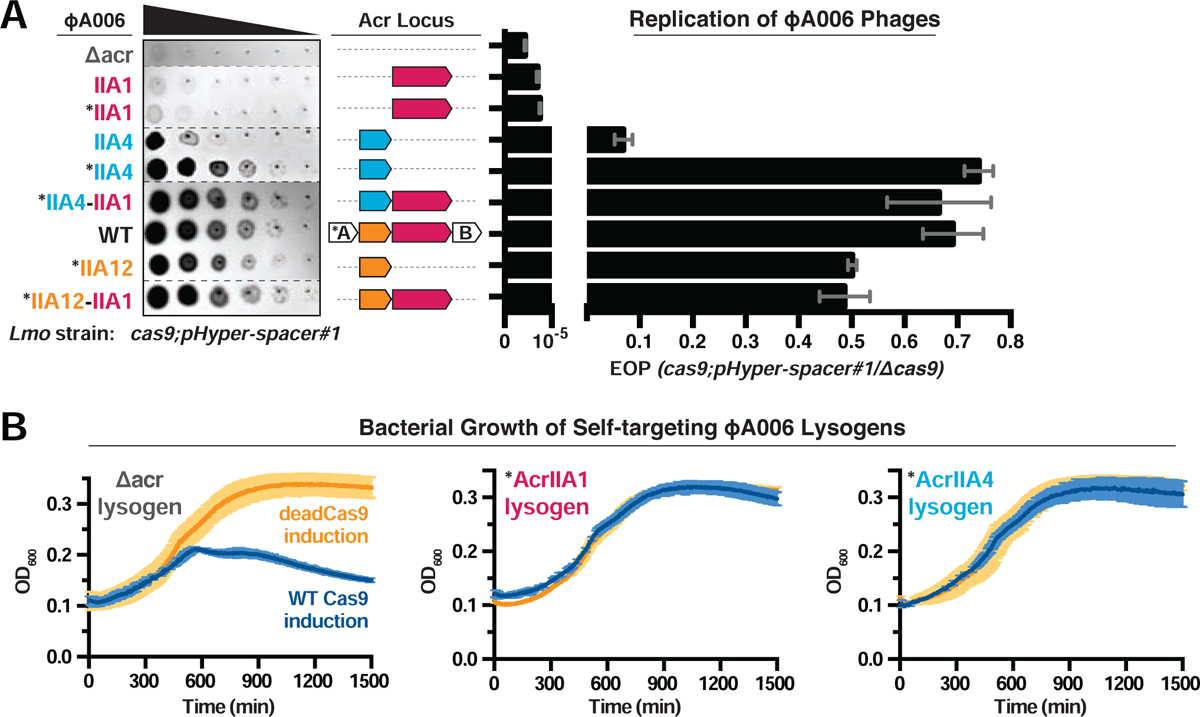
(A) Left: Representative image of plaquing assays where isogenic ΦA006 phages are titrated in ten-fold serial dilutions (black spots) on a lawn of Lmo10403s (gray background). Dashed lines indicate where intervening rows were removed for clarity. Right: Efficiency of plaquing (EOP) of isogenic ΦA006 phages expressing the indicated Acrs on Lmo10403s. Plaque forming units (PFUs) were quantified on Lmo10403s overexpressing the first spacer in the native CRISPR array that targets ΦA006 (cas9;pHyper-spacer#1) and normalized to the number of PFUs measured on a non-targeting Lmo10403s-derived strain (Δcas9). Data are displayed as the mean EOP of at least three biological replicates ± SD (error bars). See Figure S3A for EOP measurements of additional ΦA006 phages. (B) Bacterial growth curves of self-targeting Lmo10403s::ΦA006 isogenic lysogens expressing the indicated Acrs and rhamnose-inducible WT or dead LmoCas9. WT LmoCas9 (blue lines) is lethal in an Acr-deficient (Δacr) strain because the Lmo10403s CRISPR array contains a spacer targeting the ΦA006 prophage integrated in the bacterial genome. Data are displayed as the mean OD600 of at least three biological replicates ± SD (error bars) as a function of time (min). Asterisk (*) indicates the native orfA RBS (strong) in ΦA006 was used for Acr expression.
