Figure 5. AcrIIA1 is a Broad-Spectrum Cas9 Inhibitor.
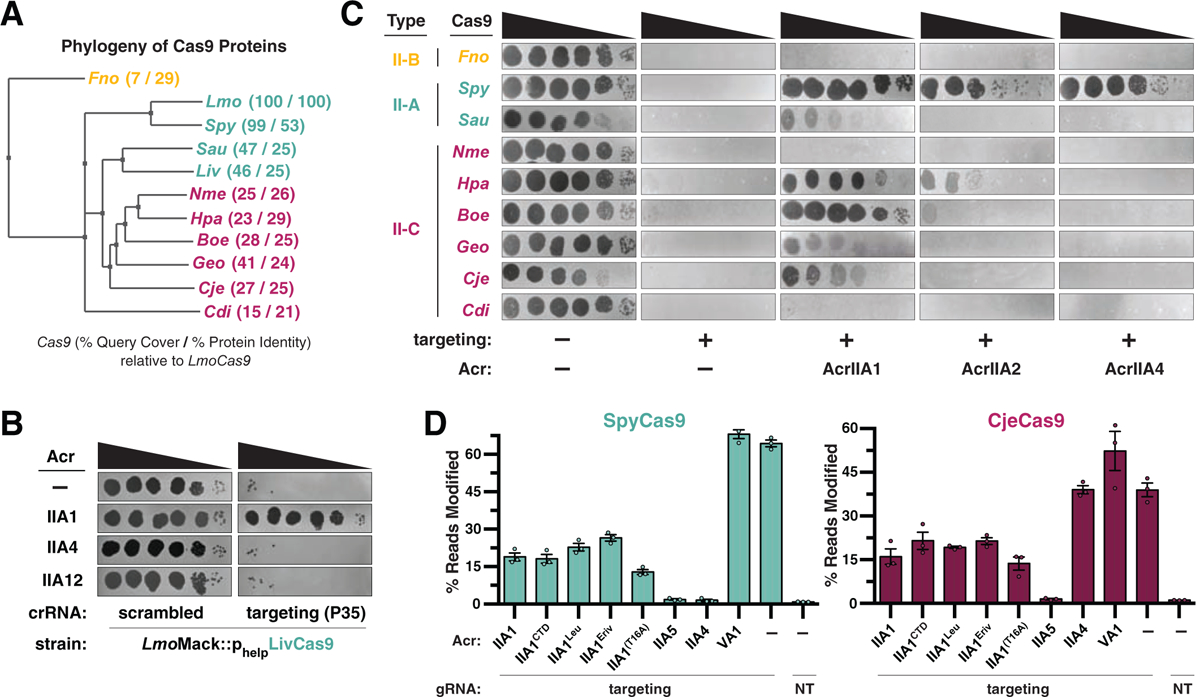
(A) Phylogenetic tree of the protein sequences of Cas9 orthologues. The percent query coverage and percent protein sequence identities relative to LmoCas9 are listed in parentheses. Cas9 orthologue names: Francisella novicida (Fno), Listeria monocytogenes (Lmo), Streptococcus pyogenes (Spy), Staphylococcus aureus (Sau), Listeria ivanovii (Liv), Neisseria meningitidis (Nme), Haemophilus parainfluenzae (Hpa), Brackiella oedipodis (Boe), Geobacillus stearothermophilus (Geo), Campylobacter jejuni (Cje), Corynebacterium diphtheriae (Cdi). (B) Plaquing assays where the Listeria phage ΦP35 is titrated in ten-fold serial dilutions (black spots) on lawns of L. monocytogenes Mack (gray background) strains that express chromosomally-integrated LivCas9/tracrRNA and contain pLEB plasmids expressing two components: an Acr or no Acr (−) and a crRNA that targets phage DNA or a scrambled crRNA (non-targeting control). (C) Plaquing assays where the E. coli phage Mu is titrated in ten-fold serial dilutions (black spots) on lawns of E. coli (gray background) expressing the indicated anti-CRISPR proteins and Type II-A, II-B and II-C Cas9-sgRNA programmed to target phage DNA. Representative pictures of at least 3 biological replicates are shown. (D) Gene editing activities of SpyCas9 and CjeCas9 in human cells in the presence of AcrIIA1 variants and orthologues. Control inhibitors (references in Methods): AcrIIA4 selective inhibitor of SpyCas9; AcrIIA5 broad-spectrum Cas9 inhibitor; AcrVA1 Cas12 inhibitor (negative control for Cas9 orthologues). Editing assessed by targeted sequencing. NT indicates a no-sgRNA control condition. Error bars indicate SEM for three independent biological replicates. See Figure S5C for editing experiments with additional Cas9 orthologues.
