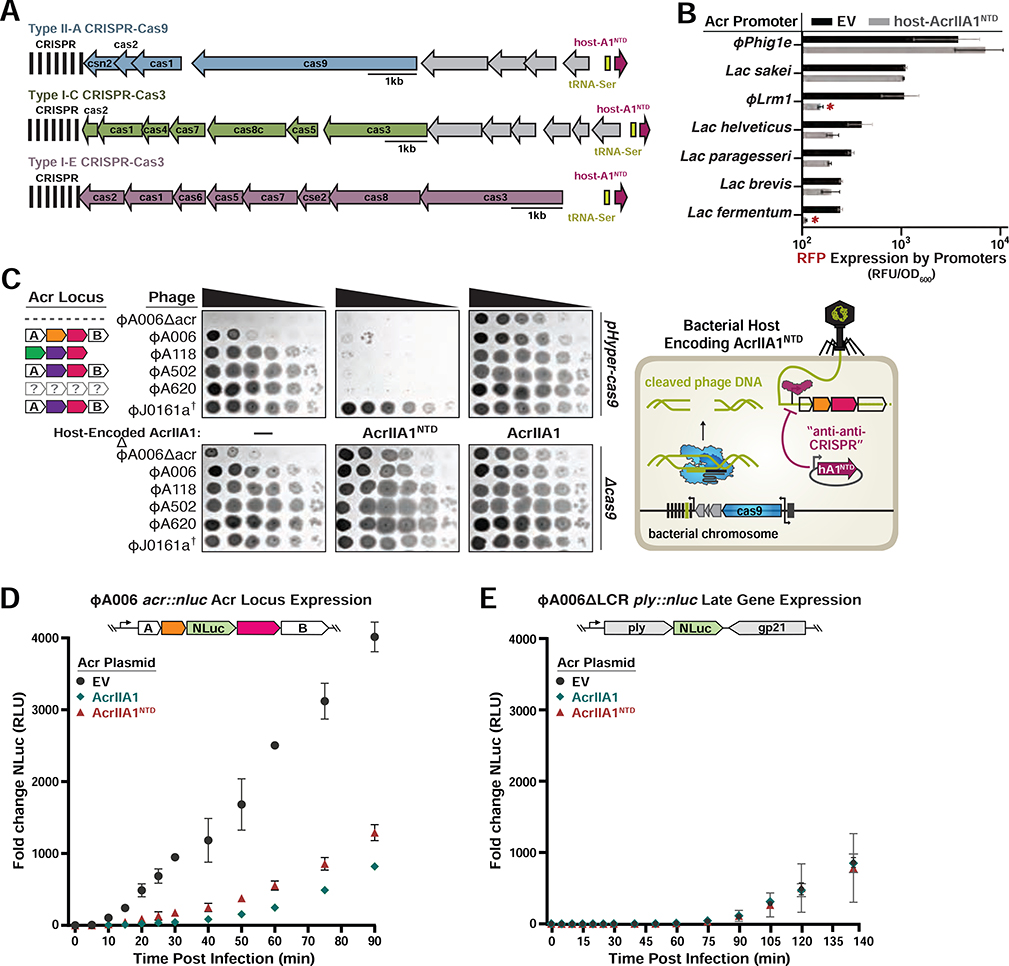
Figure 4. AcrIIA1NTD Encoded from a Bacterial Host Displays “anti-anti-CRISPR” Activity.
(A) Schematic of host-AcrIIA1NTD homologs encoded in core bacterial genomes next to Type II-A, I-C, and I-E CRISPR-Cas loci in Lactobacillus delbrueckii strains. (B) Seven promoters from the indicated phages and prophages were placed upstream of RFP, in the presence or absence of host-encoded AcrIIA1NTD, and fluorescence readout as in Figure 3. (C) Left panels: Plaquing assays where the indicated L. monocytogenes phages are titrated in ten-fold dilutions (black spots) on lawns of L. monocytogenes (gray background) expressing anti-CRISPRs from plasmids, LmoCas9 from a strong promoter (pHyper-cas9) or lacking Cas9 (Δcas), and the natural CRISPR array containing spacers with complete or partial matches to the DNA of each phage. (†) Denotes the absence of a spacer targeting the ΦJ0161a phage. Representative pictures of at least 3 biological replicates are shown. Right panel: Schematic of bacterial “anti-anti-CRISPR” activity where host-encoded AcrIIA1NTD (hA1NTD) blocks the expression of anti-CRISPRs from an infecting phage. (D) Nanoluciferase (NLuc) expression from the anti-CRISPR locus promoter or a (E) late viral promoter during lytic infection (Meile et al., 2020). L. monocytogenes 10403S strains expressing AcrIIA1 or AcrIIA1NTD from a plasmid were infected with reporter phages ΦA006acr::nluc or ΦA006 ΔLCR ply::nluc. Data are shown as the mean fold change in RLU (relative luminescence units) of three biological replicates ± SD (error bars).