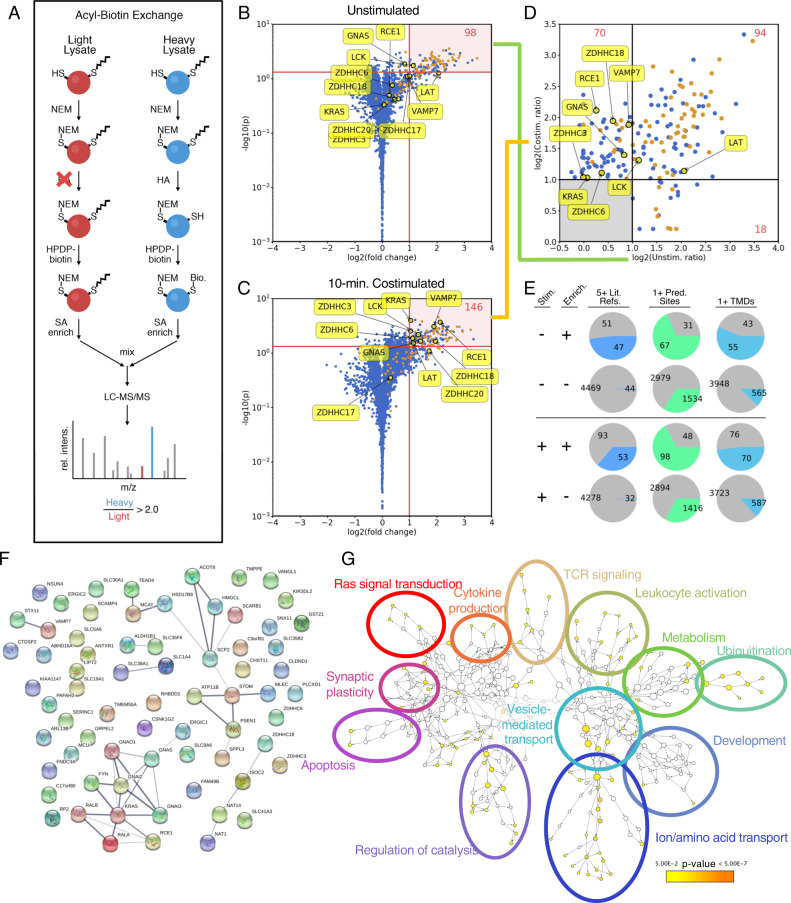
Fig. 1. Enrichment of Jurkat T-cell palmitome before and after anti-CD28/anti-CD3-costimulation by acyl-biotin exchange.
a Schematic representation of acyl-biotin exchange (ABE) protocol coupled with SILAC for quantitation of palmitoylated proteins via LC-MS/MS. Briefly, Jurkat cells grown in heavy SILAC media (blue) were lysed and free thiols were blocked with NEM. After cleaving palmitoyl groups with HA, previously palmitoylated thiols are biotinylated with HPDP-biotin, allowing for streptavidin enrichment. For control cells grown in light SILAC media (red), HA is omitted, preventing biotinylation. Proteins quantified with heavy/light intensity ratios >2.0 are considered enriched and palmitoylated. b Volcano plot showing high-confidence palmitoylated proteins prior to stimulation (n = 6, p < 0.05). Proteins found in five or more other palmitome studies are highlighted in orange. c Volcano plot showing high-confidence palmitoylated proteins 10 min after anti-CD28/anti-CD3 costimulation (n = 4, p < 0.05). d Comparison of high-confidence palmitoylated pools before and after stimulation. Only proteins from b and c that displayed a p-value < 0.05 are considered. These p-values and additional statistical parameters are detailed in Supplementary Data 1. e Analysis of palmitoylated proteins compared to unenriched (background) proteins, with three metrics: at least five references in previous palmitome studies, at least one predicted palmitoylation site (CSSPalm 4.0, high cutoff), and at least one predicted transmembrane domain (TMHMM 2.0). f STRING functional analysis of proteins enriched for palmitoylation 10 min after TCR stimulation. A clear cluster of Gα and Ras-related proteins is observed. Several interesting candidates are highlighted, including VAMP7 and DHHC18, which were investigated in more detail in this study. g Gene Ontology (GO) term enrichment of stimulation-dependent palmitoylation candidates, generated using BiNGO. Cluster size indicates the number of genes in each node, while color indicates statistical significance of enrichment.