Abstract
Helicobacter pylori is a gram-negative, spiral-shaped bacterial pathogen and the causative agent for gastritis, peptic ulcer disease and classified as a WHO class I carcinogen. While the prevalence of H. pylori infections in Africa is among the highest in the world, the incidence of gastric cancer is comparably low. Little is known about other symptoms related to the H. pylori infection in Africa and the association with certain phenotypes of bacterial virulence. We established a network of study sites in Nigeria (NG) and South Africa (ZA) to gain an overview on the epidemiological situation. In total 220 isolates from 114 patients were analyzed and 118 different patient isolates examined for the presence of the virulence factors cagA, vacA, dupA, their phylogenetic origin and their resistance against the commonly used antibiotics amoxicillin, clarithromycin, metronidazole and tetracycline. We report that H. pylori isolates from Nigeria and South Africa differ significantly in their phylogenetic profiles and in their expression of virulence factors. VacA mosaicism is intensive, resulting in m1-m2 vacA chimeras and frequent s1m1 and s1m2 vacA subtypes in hpAfrica2 strains. Gastric lesions were diagnosed more frequent in Nigerian versus South African patients and H. pylori isolates that are resistant against one or multiple antibiotics occur frequently in both countries.
Subject terms: Biochemistry, Cell biology, Microbiology, Cancer, Gastrointestinal diseases, Infectious diseases
Introduction
H. pylori is a gram-negative, spiral-shaped, highly motile bacterium that colonizes the stomach of 50–80% of people around the globe1,2. It is acquired during early childhood and usually persists for lifetime3. Prevalence varies between industrial and developing countries and is influenced by multiple factors as age, host genetic predisposition, sanitation, dietary and socioeconomic factors1,4. While the infection remains asymptomatic in most infected people, it can progress to gastritis, peptic ulcer disease and furthermore is a risk factor for the development of gastric cancer and MALT lymphoma5,6.
It has been reported that a substantial discrepancy exists between the H. pylori prevalence and gastric cancer incidence in sub-Saharan Africa, termed the “African enigma”7. Similar phenomena have also been described for other countries with a often lower state of development as India, Bangladesh and Pakistan or even Brasil8,9. While the prevalence for H. pylori infection in such countries is exceptionally high (estimated prevalence for Nigeria 87.7% and for South Africa 77.6%2, the rate of complications is usually comparably low.
In addition to host susceptibility or environmental and dietary impacts, multiple bacterial virulence factors have been studied in the context of disease progression. The most important is the cytotoxin-associated-gene pathogenicity island (cagPAI) that harbors a type IV secretion system (T4SS) that injects the effector protein CagA into gastric epithelial cells, where it is phosphorylated by Src kinases at cognate EPIYA motifs10,11. Certain forms of EPIYA motifs and the presence of cagA itself, have been reported to be associated with a higher pathology12–14.
Apart from CagA, also the vacuolating cytotoxin A (VacA) exhibits cytotoxic and immunomodulatory functions15,16. It is a pore-forming toxin, which induces the formation of vacuoles in gastric epithelial cells and can cause apoptosis17,18. The toxin is about 88 kDa in size and is structured into a signal (s), intermediate (i) and middle (m) region19. Different vacA variants have been statistically associated with pathologic features; for example, the variant s1m1 has been linked to a more severe pathology14,20. While almost all isolates from West Africa or East Asia express the s1m1 variant, the more archetypical type-2 isolates are known to lack the cagA gene and express an s2m2 vacA gene21,22.
A virulence factor that was specially linked to gastric ulceration is the duodencal ulcer promoting gene (dupA). Since the gene was first described in 2005 multiple studies with contradictory results about the role of dupA in pathogenesis have been published23–27. The dupA gene is part of a type IV secretion system located on an integrating conjugative element (ICEHptfs4). It has been previously shown that isolates of West African origin have a specific composition with an ICEHptfs4a-variant at the left junction and an ICEHptfs4b-variant homolog at the right junction (ICEHptfs4a/b, also recently designated as L2C1R128). A high proportion of West African isolates also showed a truncation close to the right junction(L2C1R1f)28, that was not found in isolates from European origin29.
Antibiotic resistance is a rising problem, causing eradication therapy failure for H. pylori worldwide. The problem is especially high in countries with limited access to a professional health care system and prescription-free sale of antimicrobial substances. In Nigeria and South Africa two therapy regimens are mainly used: a triple therapy, consisting of a proton pump inhibitor and two antibiotics (amoxicillin and clarithromycin, or metronidazole and clarithromycin) or a quadruple therapy that includes a proton pump inhibitor, bismuth and two antibiotics (amoxicillin and clarithromycin, or metronidazole and tetracycline)6. There is an urgent need to adjust standard therapy protocols according to the resistance situation. We therefore tested H. pylori isolates for resistance against the four commonly used antibiotics: amoxicillin, clarithromycin, metronidazole and tetracycline.
This work aims to gain more insight into H. pylori infections in Nigeria and South Africa, the virulence pattern of the respective isolates and their resistance to commonly used antibiotics. Therefore, we established a network of study sites and cooperation partners in Nigeria and South Africa. Isolates, patient questionnaires, gastroenterological and histopathological diagnosis from 1121 H. pylori participating patients have been evaluated, resulting in an extended overview about H. pylori infections in West- and South Africa.
Results
Study design and demographic data
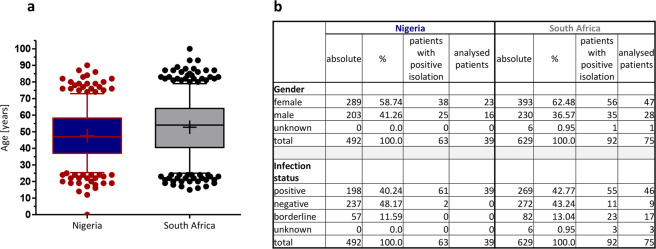
Throughout the course of this study, 492/629 (NG/ZA) patient questionnaires, 486/622 forms with gastroenterological results and 88/132 isolates (from 39/75 patients) were obtained for further analysis, the workflow of which is summarized in Fig. 1. The age distribution of the Nigerian and South African cohorts and demographic data of all patients contributing to the study are summarized in Fig. 2a,b. Patients with either a positive result in UBT (test) or a positive PCR result in antrum and corpus biopsy were considered to be H. pylori positive. Whereas the UBT test was considered as the first-line H. pylori diagnostics tool, the use of a different method was necessary in South Africa (PCR detection of H pylori DNA) due to the restricted access of the UBT kits during the sampling phase. We compared, however, the accuracy and reliability of the two methods for 113 patients, for which both test results (UBT and PCR) were available. Compared to the UBT test, no positive patients remained undetected by PCR, but 2.65% of patients with negative UBT tests were detected to be H. pylori positive (positive PCR result in antrum and corpus). From these 113 patients 10 patients (8.85%) with negative UBT result showed a positive PCR result in either antrum or corpus and were therefore classified as borderline positive (Fig. 2b). We therefore considered the PCR detection of H. pylori for South African patients as coequal to the UBT method.
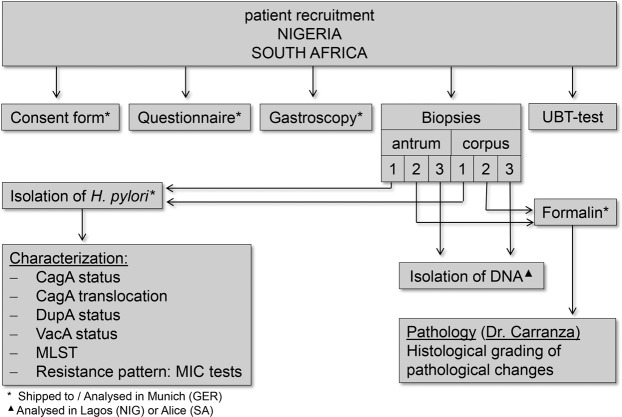
Figure 1.
Schematic overview of the study design. Patients were recruited in the Gastroenterology departments of eight different hospitals in Nigeria and South Africa. During gastroscopy diagnosis was noted and biopsies taken. From the biopsies H. pylori was cultured, DNA isolated and one biopsy of antrum and corpus fixed for pathohistological analysis.
Figure 2.
Demographic data of participating patients. (a) Depicted are box and whisker plots (5–95% percentile) for the age distribution of the study population at the time of gastroscopy. (b) The table shows the demographic distribution of gender and infection status within the study population. Infection status was determined via UBT in Nigeria and with 16 s rRNA PCR in South Africa.
We found 2/11 (NG/ZA) patients with negative diagnosis and 0/23 (NG/ZA) patients with a borderline disease status, but successful H. pylori reisolation. For statistics that included the full dataset of all patients, these patients were considered as infected, while all other patients with borderline disease status were excluded from the analysis. The mean age at the time of gastroscopy was 47.79 ± 14.71 in Nigeria and 52.69 ± 16.37 in South Africa (Fig. 2b).
Endoscopic observations and histopathological results show different patterns of disease in Nigeria and South Africa
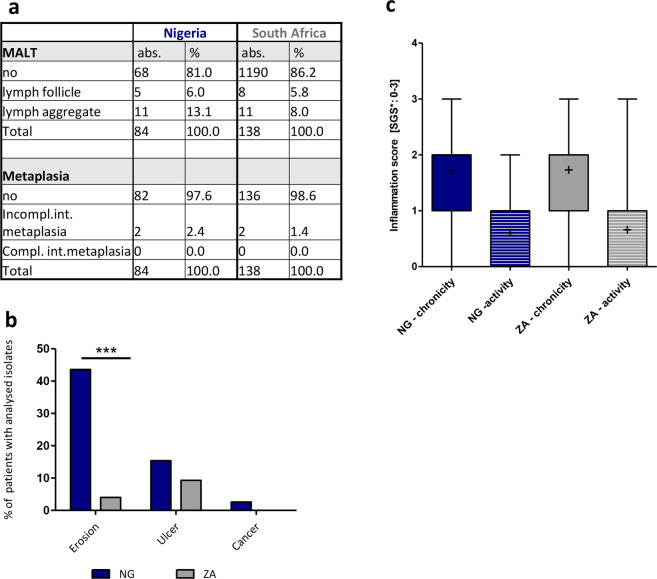
As described above, the gastroenterological examination found moderate numbers of patients presenting with symptoms of chronic gastritis. Generally, the rate of gastric complications was slightly higher in Nigeria than in South Africa, as determined by histopathological examinations (Fig. 3a). By endoscopy erosion were the most frequent diagnosis for patients in Nigeria (NG: 43.6%; ZA: 4%) and notably significantly higher in Nigerian than in South African patients (Fig. 3b). Furthermore, gastroenterological diagnosis identified gastric ulcers in both countries (NG: 15.4%; ZA: 9.3%), whereas cancer cases were reported in Nigeria only (2.6%) (Fig. 3b). However, they have not been verified by histopathology due to restrictions in the number of biopsies. There was no statistical association between the diagnosis of erosion or ulcer in South African patients with the phylogenetic profile of the patient’s isolates (e.g. hpAfrica1 or hpAfrica2), or with the ability of the respective isolate to inject CagA into the host cell. When we compared the H. pylori positive study population to the H. pylori negative participants we found that among the latter patients erosions were also most frequent (NG: 35.4%; ZA: 5.1%), followed by ulcers (NG: 16.5%; ZA: 12.1%). Cancer cases were rare (NG: 0.8%) or absent (ZA). There was no association between the intake of antibiotics, proton pump inhibitors and NSAID with the presence of ulcers, erosions or cancer. This association was tested for all patients regardless of their H. pylori infection status and for the combination of positive H. pylori infection plus medication intake (data not shown).
Figure 3.
Comparison of gastroenterological and histopathological diagnosis of patients from Nigeria and South Africa. (a) The table shows the occurrence of different forms of MALT (mucosa associated lymphoid tissue) and types of metaplasia, as determined from gastric antrum and/or corpus sections (84 NG/138 ZA) obtained from 38 patients (NG) and 74 patients (ZA) by histopathology. The updated Sidney grading system (SGS)53 was used for classification. Incomp. = incomplete; comp. = complete; int. = intestinal. (b) Percentages of patients with the diagnosis erosion, ulcer or cancer within the patient cohort. The diagnosis was given by the gastroenterologists in Nigeria and South Africa, but could not be confirmed by histology due to a limited number of biopsies. (c) The figure shows box and whisker plots (5–95% percentiles) for the presence of active or chronic inflammation. Inflammation was scored according to the SGS with a range from 0 to 3. The mean value is marked by a cross.
For further analysis we subjected stomach biopsies from the patients with positive H. pylori isolation to pathohistological examination. Due to autolysis or missing biopsies in Nigeria 84 biopsies from 38 patients and 138 biopsies from 74 patients in South Africa were analyzed. From each included patient at least one biopsy from the antrum and one from the corpus part of the stomach were available. As also reported earlier22, the disease outcome was very mild, but unexpectedly, there was no significant difference between patients from Nigeria and South Africa, which was surprising. In our cohorts of both countries, no patients with atrophy, dysplasia, cancer, erosion or ulcers were detected histologically. There was also no significant difference in activity or chronicity of disease between Nigerian and South African patients (Fig. 3c).
Patients from Nigeria and South Africa harbor H. pylori with different phylogenetic profiles
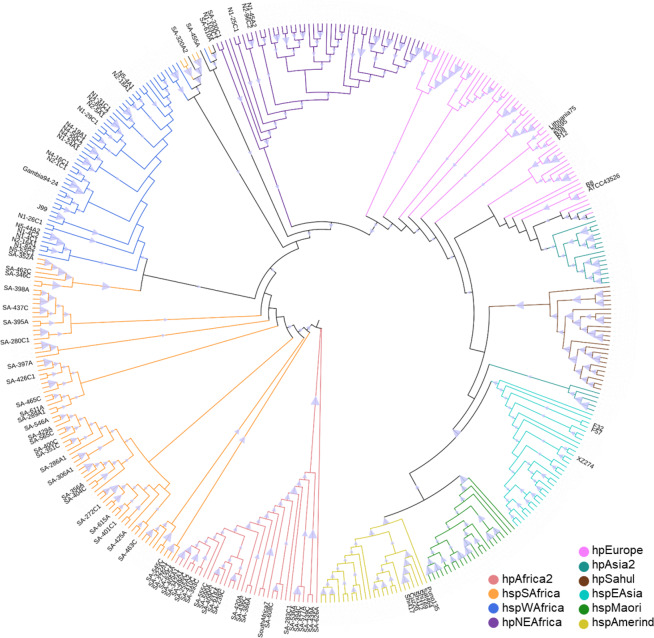
Knowing from previous studies that H. pylori isolates from the southern regions of Nigeria are mainly of hspWestAfrica subtype22, and therefore hpAfrica1, we analyzed 20 representative isolates. Except for four patients, whose isolates were of hpNEAfrica origin, all isolates clustered into the expected hspWestAfrica group, as determined by multilocus sequence typing (MLST) (Fig. 4). One of the patients with an isolate originating from Northeast Africa had a history of living in Niger and Mali. Thus, the phylogenetic profiles of Nigerian strains behaved as expected and described before. We next analyzed 56 isolates from South African patients comprising an ethnically more heterogeneous study population as compared to the Nigerian patients and little was known about the phylogeny of H. pylori in South Africa. Of these 56 isolates tested, 24 were PCR-negative for the presence of the cagA-gene, suggesting they are Type-2 strains21 whereas 32 isolates carried a cagA gene. As expected, most of the isolates lacking the cagA gene fell into the group of hpAfrica2 strains (68.8%), while the rest clustered among hpAfrica1 strains (Fig. 4). Of the 56 isolates, 22 (39.3%) clustered into the hpAfrica2 branch, 30 (53.6%) represent hspSouthAfrica and 1 (1.8%) hpNEAfrica. Three isolates (5.3°%) (SA-610A, SA-455A, SA-322A2) clustered into a branch that is clearly hpAfrica1, but includes traits from hspWestAfrica and hspSouthAfrica (Fig. 4) and therefore we were not able to classify these isolates into a distinct subpopulation.
Figure 4.
Phylogenetic tree of the isolated H. pylori strains. The evolutionary history was computed using the Neighbor-Joining method with 500 bootstrap replicates. The evolutionary distances were computed using the Kimura-2 method. This analysis involved 408 nucleotide sequences. Evolutionary analyses were conducted in MEGA X48 (https://www.megasoftware.net/). References were taken from the Helicobacter pylori MLST Databases. Reference strains are not annotated but colour-coded. The triangle displays bootstrap values from 0 to 1 drawn to scale.
Virulence factors vary significantly between H. pylori isolates from Nigeria and South Africa
As mentioned in “demographic data” more than one isolate per patient was analyzed, but not all isolates could be successfully transferred to the analyzing lab in Munich, limiting the study of the virulence factors to isolates from 39/75 patients, respectively. To avoid statistical bias by including the same isolates twice, we evaluated all isolates with RAPD analysis. Except for three patients, all patients harbored RAPD-identical strains. When the isolates were identical, only one isolate per patient was included. Three patients harbored non-identical strains, therefore all RAPD non-identical strains were included, resulting in 41/77 isolates to be analyzed.
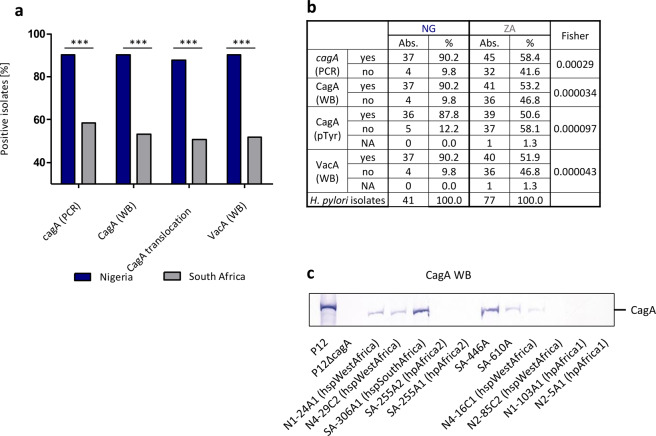
We evaluated the isolated strains by PCR and Western blotting for the presence of cagA and vacA (Fig. 5a,b) and also performed functional assays for the translocation and phosphorylation of CagA into host cells. The frequent presence of hpAfrica2 subtypes in South Africa resulted in a significant difference in expression of vacA and cagA. While all isolates of hpAfrica2 origin were negative for cagA, the vacA subtypes showed a mixed phenotype (Figs. 5a,b, 6c).
Figure 5.
The expression of virulence factors CagA and VacA differs between Nigerian and South African isolates. (a) Percentages of patient isolates positive for the presence of cagA (as determined by PCR), the expression of CagA and VacA (as determined by western blotting (WB)) or functional CagA translocation as determined by tyrosine phosphorylation assay (pTyr). Groups were compared using Fisher’s exact tests. *p < 0.05, **p < 0.01, ***p < 0.001. (b) Statistical analysis of CagA and VacA results. (c) Western blot showing the presence or absence of CagA protein in a set of Nigerian or South African H. pylori isolates compared to H. pylori P12 reference strain.
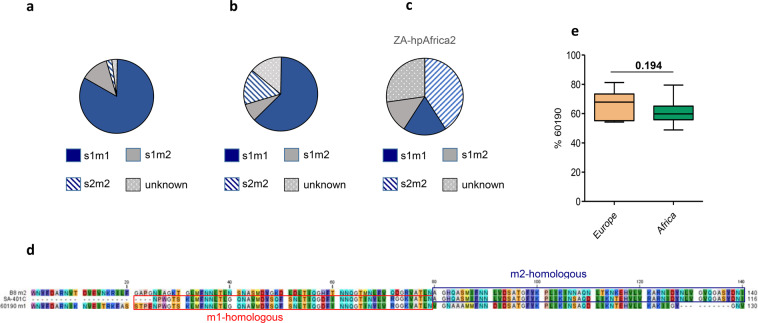
Figure 6.
VacA mosaicism differs significantly between Nigerian and South African H. pylori isolates. (a) vacA mosaicism of isolates obtained from Nigerian patients. (b) vacA mosaicism of isolates from South Africa. (c) Representation of vacA mosaicism of a sub-cohort of hpAfrica2 isolates from South Africa. (d) Depicted is the amino acid sequence in the mid-region of the isolate SA-401C and the m1-reference sequence of 60190 (red box) and the m2-reference sequence of B8 (blue box) demonstrating that SA-401C is a chimera. (e) Depicted is the extent of induced cell vacuoles in HeLa cells relative to H. pylori control strain 60190. Compared are 11 strains isolated from Europe with 6 isolates from Nigeria and 4 isolates from South Africa in three independent experiments. All isolates from Africa were of vacA subtype s1m1. P12, 60190 and P12∆vacA served as controls.
Interestingly, four of 77 isolates from South Africa were positive for cagA PCR, but did not express CagA as detected by Western Blot (SA-289A1, SA-330C, SA-378A; SA-480C). Sequencing cagA we found frameshift mutations at different sites in SA-289A1, SA-330C and a SNP leading to a stop codon in SA-480C. Three hpAfrica1 isolates lacked the cagPAI, as determined by Western blotting and sequencing (N2-85C2; N1-103A1C1; N2-5A1) (Fig. 5c), whereas eight isolates from South Africa, which were all hpAfrica2, also lacked the complete cagPAI. Two isolates from South Africa and one from Nigeria were cagA PCR and WB positive, but not able to functionally inject CagA into the host cell as determined via AGS tyrosine phosphorylation assay (SA-446A, SA-610A, N4-16C1). Sequencing of cagA in these isolates was inconspicuous, suggesting a regulatory mechanism, or mutations in other parts of the cagPAI (data not shown).
Another virulence-associated factor implicated in the formation of duodenal ulcers is the dupA locus. H. pylori solates from WestAfrican origin carry this locus on an integrating conjugative element of the type ICEHptfs4a/b29. It was previously shown that this element lacks the 5′-region of dupA in isolates of West African origin, because of a truncation, which is shortening the island by around 3.8 kb at the right junction29. To compare between the cohorts, we utilized PCR analysis and amplified a DNA fragment spanning this truncation, or the 5′ region of non-truncated dupA. The 3.8 kb truncation was present in 82.4% of isolates from Nigeria, but only in 50.7% of isolates from South Africa. Comparing this result with the phylogenetic heritage of the isolates revealed that all isolates with a truncated ICEHptfs4 were of hpAfrica1 subtype. Of the analyzed 22 hpAfrica2 isolates, 16 had a full-length dupA gene, and in 6 isolates we could not amplify this region. Furthermore, we detected 6 isolates that showed evidence of both dupA forms, thus indicating the presence of more than one integrating conjugative element, or at least parts of it.
Intensive VacA mosaicism in African H. pylori isolates and m1-m2 vacA chimeras
In order to judge the role of vacA and its contribution to virulence, we further evaluated vacA mosaicisms in the obtained isolates. Sequence variations in vacA gene mosaics differed substantially between the isolates from Nigeria and South Africa (p = 0.01303), most likely attributable to the presence of hpAfrica2 isolates in South Africa (Fig. 6a,b). The majority of Nigerian isolates (82.9%) harbored an s1m1 region, whereas the s1m2 (12.2%) and the s2m2 sequences (2.4%) were less frequent. In South African strains the s1m1 form was dominant as well (62.3%), albeit lower than for Nigerian strains, followed by s2m2 (15.6%) and s1m2, (7.8%) sequences. Notably, a separate analysis of the hpAfrica2 isolates (22) revealed the highest number of s2m2 genotypes (40.9%), while s1/m1 (18.2%) and s1/m2 (13.6%) were less abundant (Fig. 6c). Furthermore, one isolate, SA-401C, represented a chimera of m1 and m2 vacA regions, as recently described30 (Fig. 6d). An s1b subtype (92.7% versus 88.9%, respectively) dominated in Nigerian as well as in South African strains the s-region subtypes. Remarkably, from 11 South African isolates (14.3%) it was not possible to determine the subtype of either m- or s-region.
We next analyzed the vacuolating activity of both strain cohorts using a quantitative vacuolization assays as previously described31,32 and compared cytotoxic activity of African strains with European isolates. Ten randomly chosen isolates from Africa (s1m1 phenotype) were compared to 11 strains of European origin. As a result, the median vacuolating cytotoxicity of the African isolates was slightly lower as compared to the European isolates, but the difference was not significant (Fig. 6e).
Antibiotic resistance as a common problem in Nigeria and South Africa
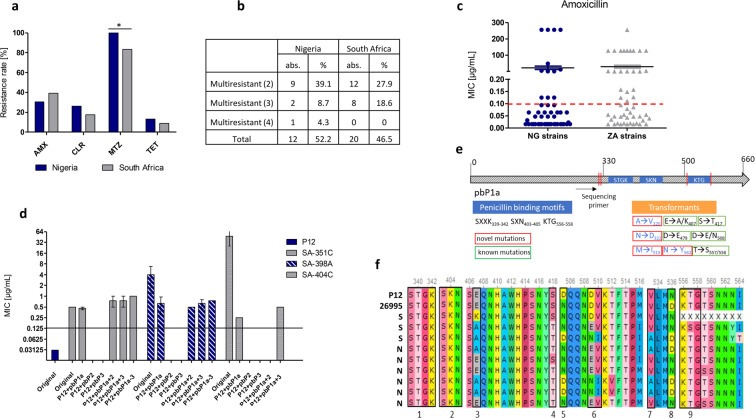
Antibiotic resistance is a known problem in developing countries with high disease burden and wide access to antibiotics (over-the-counter-policy). We evaluated resistance rates of Nigerian and South African H. pylori isolates for commonly used antibiotic drugs amoxicillin, clarithromycin, metronidazole and tetracycline by E-test. Resistance rates varied between substance classes with the highest rate for metronidazole (NG: 100%; ZA: 83.3%) and the lowest for tetracycline (NG: 13%; ZA: 8.7%) respectively (Fig. 7a). For Nigeria 52.5% of isolates were resistant against more than one antibiotic substance with 4.3% being resistant against all four tested antibiotics (Fig. 7b). In South Africa, we found 46.5% of isolates being resistant against more than one antimicrobial substance, but all isolates were susceptible to at least one of the tested drugs. Patients with previous treatment with antibiotics (5/23 patients in Nigeria; 1/46 patients in South Africa) did not show higher resistance rates against one or multiple antibiotics.
Figure 7.
Antibiotic resistance in African isolates. (a) Rates of resistance against commonly used antibiotics amoxicillin (AMX), clarithromycin (CLR), metronidazole (MTZ) and tetracycline (TET). 23 patient isolates from Nigeria and 46 patient isolates from South Africa were evaluated. (b) Summary of isolates with multiple resistances. (c) Distribution of MIC values against amoxicillin of independent H. pylori isolates as determined by E-test. (d) MIC values of amoxicillin-sensitive P12 strain transformed with various PCR amplification products of pbp1a, pbp2 and pbp3 and their combinations generated from genomic DNA of amoxicillin-resistant African isolates. Displayed are the mean MIC values of three independent transformation experiments. MIC values were evaluated in two independent two-fold dilution growth curve experiments. The black vertical line marks the cut-off value for resistance (>0.125 mg/L). Empty bar mean that no transformants were obtained in the experiments. (e) Schematic representation of pbp1 with location of penicillin binding motifs and the positions of mutations obtained from amoxicillin-resistant transformants. Novel so far not described mutations are boxed in red, already known and published mutations are boxed in green. (f) Alterations in sequences of pbp1a in amoxicillin-resistant isolates from Nigeria (N) and South Africa (S) as compared to P12 and 26695 reference strains. Positions 1, 2 and 9 indicated below the sequences mark the penicillin binding motifs.
Low and high level of amoxicillin resistance, alterations in penicillin binding proteins, but no β-lactamase activity
After determining surprisingly high resistance rates against amoxicillin with high MIC values, we tried to evaluate possible mechanisms. An overview showing the MIC levels of individual H. pylori isolates against amoxicillin from the tested strains from Nigeria and South Africa showed a similar distribution of low-resistance and high-resistance strains in both countries (Fig. 7c). We amplified the genes for penicillin binding proteins 1a, 2 and 3 and transformed corresponding PCR products singularly and in all possible combinations into the sensitive H. pylori strain P12. To verify MIC values we determined growth curves in the presence of two-fold dilutions of amoxicillin. In general, the obtained transformants displayed only a low-level resistance against amoxicillin and did not reach resistance levels of the original patient isolates (Fig. 7d). For a successful generation of resistance transfer of pbp1a from the resistant strain was necessary, whereas other pbp genes, or other unrelated control DNA fragments did not result in an amoxicillin resistant phenotype. We sequenced pbp1a of representative transformed P12 isolates and found multiple alterations, but no unifying feature for all resistant isolates (Fig. 7e,f), suggesting that low-level amoxicillin resistance is generated by a cumulative effect of multiple sequence alterations in pbp1, as reported previously33–36. As postulated by other research groups H. pylori high-level amoxicillin resistance may be due to a β-lactamase encoded in blaTEM-137. To test for β-lactamase activity we investigated resistant isolates using a commercially available Nitrocefin test. None of the isolates revealed a detectable β-lactamase activity. To control for other mutations or genetic alterations not encoded in penicillin binding proteins we also transformed whole genomic DNA of highly amoxicillin-resistant clinical isolates (MIC > 256 µg/ml) into the sensitive P12 strain. However, transformants with low-level resistance were readily obtained, but no transformants displaying MIC values >2 µg/ml could be observed. In conclusion, this suggests that the mechanism for amoxicillin resistance in H. pylori is probably not encoded by a single gene, but might be the result of multiple allelic alterations.
Discussion
The prevalence of H. pylori has been declining in many highly industrialized countries of the Western world, whereas it stayed constant at a relatively high level in certain developing countries. This has major implications on the sequelae associated with an H. pylori infection, including peptic ulcer disease and gastric cancer. In this study, we intended to get more insights in H. pylori patient isolates from Nigeria versus South Africa and to compare their phylogenetic profiles, their virulence factors, and their antibiotic resistance profiles, known to be a common problem in Nigeria and South Africa. To avoid statistical bias or overestimation of certain characteristics, we decided to interpret only one H. pylori isolate per patient when the isolated strains were the same as determined by RAPD PCR analysis. This procedure resulted in a relatively low number of examined isolates. We adjusted statistic testing to the Fisher’s exact test that is suitable for small numbers of cases. All patients of the Nigeria and the South Africa cohorts are listed in Fig. 2b and differentiated according to gender and infection status (column absolute), and the cases for which H. pylori isolates could be obtained (column: patients with positive isolation). Only these latter patients and patient isolates, for which all tests have been performed, have been included into the evaluation of our results. We also assume that the set of isolates tested in detail is representative for the whole 1121 patients included in the study, since the gender and age distribution of the patient set with isolates studied is in a similar range as that of all patients. The relatively low number of isolates that could be analyzed in the lab as compared to the relatively high number of participants has multiple reasons. Due to the fastidious nature of H. pylori, isolation rates have been reported to vary greatly (30–70%) between laboratories38,39. In Nigeria it was not possible to isolate H. pylori directly at the sampling site and with difficult infrastructure conditions there were significant delays in the isolation procedure. Isolates were then shipped to Munich causing a significant proportion of isolates that could not be regrown from the transport medium due to a long transport time.
By comparing the disease outcome of Nigerian and South African patients and the patient isolates, it was obvious that the number of severe complications of H. pylori infected patients, like erosions (NG: 43.6%; ZA: 4.0%), ulcers (NG:15.4%; ZA: 9.3%) or cancer (NG: 2.6%; ZA: 0%) was lower in South African patients than in Nigerians. A low gastric cancer rate in the African countries might be explained by a generally lower life expectancy as compared to many other countries. Furthermore, it could be speculated that one reason for that might be the high number of hpAfrica2 strains in South Africa, which lack the main virulence factor, the cagPAI and are supposed to mainly express the less virulent vacA subtype s2m2. However, our data show now that more pathogenic s1m1 and s1m2 vacA subtypes (together 31%) are found in hpAfrica2 strains, which is unexpected and challenges the hypothesis that preferentially H. pylori type I strains, carrying a functional cagPAI, express the s1m1 vacA19. Our data show that vacA s1m1/m2 is well compatible with the absence of the cagPAI and suggests that in such ancient, cagPAI deficient strains both types, s1m1/m2 and s2m2 vacA genes were existent. With acquisition of the cagPAI during evolution by H. pylori a selection pressure might have favored the co-expression of s1m1 vacA subtypes with these strains.
We were not able to link the association statistically, because the case numbers with gastroenterological disease were in general quite low. The pathology examination could not confirm erosions or ulcerations and it remained unclear if this was due to sampling errors during gastroscopy (e.g. sampling in the wrong areas), or interobserver variations. However, in this context similar complications have already been described in the past22,40.
The continuous increase of antibiotic resistance levels in bacteria is a worldwide problem41,42, but cannot be overemphasized in developing countries with difficult access to professional health care and open sale of antibiotics. In our study, antibiotic intake within the past three months did not directly influence resistance, but 52.12% (NG) and 46.51% (ZA) of isolates were resistant against more than one antibiotic. Overall, the resistance rates we observed in our study were well in the range of a recent meta-analysis published by Jaka et al.43.
Notably, only 15% (NG) and 1.3% (ZA) of all patients included in this study declared to be pretreated with antibiotics in the past. All patients with positive UBT or PCR result were considered as H. pylori infected, but we included only patients with successful H. pylori isolation into the further study. To control for a bias caused by positive selection of untreated patients, we next compared all patients with positive UBT or PCR result for H. pylori. From this cohort 16.5% (NG) and 3.9% (ZA) of patients, respectively, had declared to be pretreated with antibiotics (data not shown), which is comparable to the overall patient cohort and argued against a bias in our studied patient cohort.
Since antibiotic resistance against amoxicillin has been discussed quite controversial and the reported resistance rates vary quite drastically43, we decided to study the responsible mechanisms. None of the isolates tested exhibited a β-lactamase activity, as originally reported by Tseng et al. for amoxicillin resistant H. pylori strains37. We next generated pbp1a, pbp2 and pbp3 DNA fragments from highly amoxicillin-resistant H. pylori isolates (Fig. 7e). Transformation of pbp1a, but not pbp2 or pbp3 into the sensitive P12 strain resulted in amoxicillin-resistant transformants, although the full resistance level of the original isolates was not obtained, which is different to previously reported results44. We found multiple alterations in the sequence of pbp1a, but none of the alterations was a unique feature to all resistant isolates (Fig. 7e). Interestingly the penicillin binding motifs SXXK339–342, SXN403–405 and KTG556–558 were commonly not affected (except for one isolate with a threonine to serine substitution in KTG556–558), but the region AA 500–550 in close proximity to the KTG motif was the sequence with the highest variation (Fig. 7e,f). Taken together our results suggest that single mutations in the pbp1a gene result in a low-level amoxicillin resistance of H. pylori, but other mechanism(s) conferring a high-level amoxicillin resistance are so far unknown.
Methods
Study design
Patients were recruited between November 2015 and July 2018 in different hospitals in Nigeria (NG) and South Africa (ZA). In Nigeria five participating hospitals located in Lagos, one each in Ibadan, in Ile-Ife and in Jos. Patients in South Africa were recruited from two different hospitals in Johannesburg and Port Elisabeth. Each patient filled a questionnaire and signed a consent form and a gastroenterological diagnosis was provided by the gastroenterologists in charge.
Urea breath test (UBT), which, besides a PCR or a nested PCR approach, is considered as the gold standard in H. pylori diagnostics for African countries, was performed22, and six biopsies (three from the antrum and three from the corpus) were taken from each patient. One antrum and one corpus biopsy were used for isolation of H. pylori, while the other two biopsies were used for extraction of DNA and histological analysis.
The inclusion criteria comprised the treatment in one of the mentioned hospitals and the patient agreement to participate in this study. The recruitment, the gastroenterological examination, the UBT, the fixation of the biopsies and the isolation of the strains were performed in the hospitals in Nigeria and South Africa and the corresponding laboratories of the Nigerian Institute of Medical Research, Lagos and University of Fort Hare, Alice. Throughout the course of this study, patient isolates, questionnaires and forms with gastroenterological results were transferred to the Max von Pettenkofer-Institut of the LMU in Munich, Germany. A more intensive biochemical, immunological and molecular characterization of isolates as well as a histological grading of biopsy material was performed in Munich. These assays included the determination of the status of antibiotic resistance levels (MIC tests) the presence or absence of genes encoding for H. pylori virulence factors by polymerase chain reaction (cagA, vacA), as well as the detection of expression and function of major virulence factors like CagA and VacA by western blotting, CagA translocation and vacuolization assays. A statistical analysis was performed to determine correlations between H. pylori virulence, risk factors and disease outcome.
Urea breath test
The urea breath test (UBT) was obtained from Tri-Med Distributors in Perth, Australia. UBT was performed as described previously22. Briefly, patients fasted either overnight or for a minimum of four hours. A capsule containing a known amount of 14C-labelled urea was provided to patients to drink with 30 mL of water, followed after 3 minutes by a further 30 ml of water. After further 7 minutes a breath sample is collected in a sterilized mylar balloon. Following contact between 14C-labelled urea and stomach-resident H. pylori, the molecule is hydrolyzed into 14C-carbon dioxide and ammonia. The carbon dioxide is then exhaled by the patient. The contents of the balloon were dissolved into breath collection fluid and then liquid scintillation fluid was added to quantify the degree of 14C in the sample. Scintillation values below 50 DPM (disintegrations per minute) were considered to be negative, values over 200 DPM as positive and all values between as “borderline”.
H. pylori isolation
The isolation of H. pylori from biopsy specimens was performed as described previously22. Briefly, biopsy specimens were aseptically rolled over the surface of Columbia blood agar base plates under a biological safety cabinet (Thermo Fischer Scientific). The agar (Oxoid CM0331) was supplemented with 7% horse serum (Oxoid SR0048), 1% vitamin mix (Isovitale-X), and an H. pylori selective supplement (Dent, SR0147E Oxoid) comprising of amphotericin B (2.5 mg), trimethoprim (2.5 mg), vancomycin (5.0 mg), and cefsulodin (2.5 mg). The plates were incubated at 37 °C in an atmosphere of 85% N2, 10% CO2 and 5% O2 for 4–10 days. H. pylori colonies were identified as small, round, translucent, Gram-negative bacteria and positive for catalase, oxidase and urease tests. The confirmed isolates were frozen in Brucella broth containing 20% glycerol and stored at −80 °C for future use.
Transport of H. pylori strains
For transport of the isolates from Nigeria or South Africa to Munich (Germany), strains were cultured on GC agar serum plates, passaged at least twice and transferred into Portagerm Pylori (bioMérieux SA Mercy L’Etoile France) medium for shipment at room temperature, as described previously22.
PCR and MLST
PCR was performed in 25 µl reaction mixtures, consisting of 1X PCR buffer, magnesium chloride (1.5 mM), dNTP (200 μM), primer (10 pmol) and 1U Taq DNA polymerase (Pan-Biotech GmbH). Amplification was carried out in a Peqlab Thermocycler using the following cycling parameters: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 sec, 52 °C for 30 sec and 72 °C according to amplicon length (1 min per 1000 bp). This was followed by a final extension of 72 °C for 10 min. The hpWAfrica strain J9945 and the hpEurope strain P1246 were used as control strains. Primers are listed in Table 1. For strain phylogeny, multilocus sequence typing analysis was used47. Partial sequences of 7 housekeeping genes were obtained by Sanger sequencing after purifying PCR products with Illustria GFX PCR DNA and gel band purification kit. The resulting sequences were assembled and aligned with the corresponding sequences from H. pylori reference strains from the MLST database, using the Muscle algorithm within MEGA X48. Phylogenetic trees were constructed and tested by neighbor-joining method using the Kimura 2-parameter model of nucleotide substitution, and 500 bootstrap replications.
Table 1.
Primer sequences.
| Target gene | Sense primer | Antisense primer | Amplicon |
|---|---|---|---|
| cagA | ACCGCTCGAGAACCCTAGTCGGTAATGGG | ATATCGATTTAAGCCAATTTTTGATTCCTTG | 500 bp |
| cagA (full length) | CCATCGATGG TAAAAATGTG AATCGT (454) | CAGGTACCGC GGCCGCTTAA GATTTTTGGA AACCAC (1314) | 3658 bp |
| vacA s1 | CTGCTTGAATGCGCCAAAC | ATGGAAATACAACAAACACAC | 259 bp |
| vacA s2 | CTGCTTGAATGCGCCAAAC | ATGGAAATACAACAAACACAC | 286 bp |
| vacA m1 | GGTCAAAATGCGGTCATGG | CCATTGGTACCTGTAGAAAC | 290 bp |
| vacA m2 | CATAACTAGCGCCTTGCAC | CATAACTAGCGCCTTGCAC | 352 bp |
| atpA | GGACTAGCGTTAAACGCACG | CTTGAAACCGACAAGCCCAC | 841 bp |
| efp | GGCAATTTGGATGAGCGAGCTC | CTTCACCTTTTCAAGATACTC | 559 bp |
| mutY | GTGGTTGTAGYTGGAAACTTTACAC | CTTAAGCGTGTGTYTTTCTAGG | 676 bp |
| ppa | GGAGATTGCAATGAATTTAGA | GTGGGGTTAARATCGTTAAATTG | 706 bp |
| trpC | TAGAATGCAAAAAAGCATCGCCCTC | TAAGCCCGCACACTTTATTTTCGCC | 633 bp |
| ureI | AGGTTATTCGTAAGGTGCG | GTTTAAATCCCTTAGATTGCC | 686 bp |
| yphC | CACGCCTATTTTTTTGACTAAAAAC | CATTYACCCTCCCAATGATGC | 734 bp |
| pbp1a | GATTGTCATAGGGTTGTTAGC (PP-14) | GGGTTCTTCGCTATCGTC (PP-15) | 1928 bp |
| pbp2 | GTCTTCGCTATAAGCTTTTG (PP-54) | CTCATAGAGTTTGTTGCTC (PP-55) | 1745 bp |
| pbp3 | TGATCCTTACTTCAACCCA (PP-56) | GTTTTGAATCGCAATAGAGGG (PP-57) | 1797 bp |
| dupA | CTACAATATAGCTCTCAAAAG(WS539) | AGCAATAAAACGCTTAAAAGTCTC(WS606) | 2935 bp |
| dupA | GTATTCCTAGCCAATATTCTTTAG(WS677) | AAAAATTTAGGCTCAAAGTCTG (WS678) | 597 bp |
Western blotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting was performed as described47 using polyvinylidene difluoride (PVDF) filters blocked with 3% BSA in TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl), 0.1% (v/v) Tween 20. Rabbit polyclonal antisera against CagA and VacA have been described previously49. Alkaline phosphatase-conjugated protein A or horseradish peroxidase-conjugated anti-rabbit IgG antiserum was used to visualize bound antibody.
Phosphorylation of translocated CagA
To determine capability of CagA-translocation AGS cells were infected with H. pylori strains and phosphotyrosine immunoblotting were performed10. Briefly, cells were infected with bacteria adjusted to OD550 0.2 for 4 h at 37 °C, washed three times and suspended in PBS containing 1 mM EDTA, 1 mM Na3VO4, 1 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml pepstatin. Cells were then collected by centrifugation and resuspended in sample buffer. Tyrosine-phosphorylated proteins were analyzed by immunoblotting with the phosphotyrosine antiserum PY99 (Santa Cruz Biotechnology).
Vacuolization assay
To determine cytotoxicity, a vacuolization assay, as described previously was performed32. Briefly HeLa cells were grown to 70% confluency, infected with OD550 0.1 and incubated for four hours at 37 °C and 10% CO2. After incubation with NH4Cl for another hour, cells were stained with neutral red and washed twice with PBS + 15% FCS. Cells were then incubated with 70% ethanol+ 0.37% HCl for 5 minutes and the extinction measured at 534 nm in a TECAN plate reader. Values were normalized to the control strain 60190. A H. pylori P12∆vacA mutant31 served as negative control.
MIC test
The minimal inhibitory concentration was obtained using E-test strips and performed according to manufacturer’s instructions (Liofilchem, Italy) and as described earlier22. H. pylori isolates were grown as a liquid culture in brucella broth + 10% fetal calf serum (FCS) for 3–4 hours until OD550 1.0. 400 µl of the liquid culture was transferred to a GC agar serum plate and allowed to dry for several hours. An antibiotic strip was placed on the plate. Antimicrobial agent concentrations ranged from 0.016 to 256 µg/mL. After three days incubation the zone of inhibition was measured under consideration of recommended MIC breakpoints (EUCAST Clinical Breakpoint Table v. 8.0, valid from 2018-01-01): amoxicillin >0.125 µg/mL, clarithromycin >0.5 µg/mL, tetracycline >1 µg/mL, metronidazole >8 µg/mL. Every isolate was tested twice independently with E-test.
Transformation
PCR products of penicillin binding proteins pbp1a, pbp2 and pbp3 of resistant isolates were generated (see primer listed in Table 1) and transformed into the sensitive H. pylori strain P12 by natural transformation, as described previously50. H. pylori transformants were selected on serum agar plates containing 1 mg/L amoxicillin (Sigma-Aldrich). Clones were frozen in brucella broth containing 20% glycerol and 10% FCS, stored at −80 °C and plated again on GC agar plates containing 1 mg/L amoxicillin.
Growth curves
MIC values of clones were evaluated using growth curve assays, as described earlier51. H. pylori were diluted to an OD550 0.075 and subcultured in 96-well microtiter plates (clear, flat-bottom, Costar, Corning Inc.), which were then sealed with a gas-permeable membrane (Breathe-Easy sealing membrane, Diversified Biotech). Plates were incubated at 37 °C under microaerobic conditions (while shaking) in the Clariostar plate reader (BMG Labtech). Optical density was automatically monitored and measured every five minutes. Growth curves were analyzed and processed with MARS Data Analysis software 3.10 R5 (BMG Labtech). Percental growth was expressed as [(ΔODsample/ΔODcontrol) x 100], while ΔOD corresponded to “ODstationary phase − ODlag phase”.
Nitrocefin tests
A Nitrocefin test was used to evaluate for β-lactamase activity. Nitrocefin is a chromogenic cephalosporin, with a β-lactam ring. Upon oxidation by β-lactamase color is rendered from yellow to red. Nitrocefin disks for rapid detection of β-lactamase enzymes were purchased from Sigma and used as suggested by the manufacturer. Briefly, disks were allowed to equilibrate to room temperature and were humidified with 10 µl of H2Obidest. Single colonies were streaked directly from GC agar plates on the discs and incubated for 20 minutes to observe a color change from yellow to red. H. pylori P12[TEM-1-CagA]51 served as positive control and E. coli DH5α as negative control.
Histology
Biopsies were fixed in 4% buffered formalin at the study sites and sent to Munich. Hematoxylin and eosin stained longitudinal paraffin sections of antrum and corpus (one section per location) were graded according to the updated Sydney System52 by pathologists in Munich on the intensity of inflammation, metaplasia, and presence of gastric mucosa associated lymphoid tissue (MALT), as well as the presence and occurrence of atrophies, metaplasia, dysplasia, cancer, erosion and ulcers.
Ethics
The study was reviewed and approved by the Ethics Committee of the Nigerian Institute of Medical Research (registration number IORG0002656), as well as the Ethical Committee of the Chris Hani Baragwanath Academic Hospital (CHBAH) Soweto Johannesburg (registration number M160228), the University of Fort Hare Alice Eastern Cape South Africa (registration number Rec-270710-028-RA level 01) and the Ludwig-Maximilians-University Munich (registration number 335-08). The study was conducted in line with the Declaration of Helsinki and informed consent was obtained from all study participants.
Statistical methods
Interrelations between clinical, pathologic, and laboratory findings were investigated bivariately using chi-squared or fishers exact tests (frequencies less than 5 for a statistical event)22. Due to the exploratory character of these analyses, all tests were performed on an alpha level of 5% without any correction for multiple testing. P-values depicted in this manuscript value: *p < 0.05, **p < 0.01, ***p < 0.001. All analyses were done with R: A Language and Environment for Statistical (Vienna, Austria).
Acknowledgements
The Deutsche Forschungsgemeinschaft (DFG) funded the project in the context of the German African Infectiology Initiative (HA2697/18-1) to Rainer Haas, Anna Clarke, and Stella Smith. Authors are grateful to physicians and nurses in the participating hospitals in Nigeria and South Africa for their involvement and cooperation. We would also like to appreciate excellent technical support from Evelyn Weiss, as well as administrative support from Friederike Aicher. Thanks to all patients who volunteered to participate in our study.
Author contributions
Conceptualization: R.H., S.S., A.C., U.H. Data curation: P.P., T.J., A.I., E.L., M.C., R.A., H.N. Formal analysis: P.P. Funding acquisition: R.H. S.S. A.C., U.H. Methodology: P.P., W.F., R.H., S.S., A.C., C.O., R.U., I.A., O.L., D.N., O.A., I.A.A. Supervision: R.H. Visualization: P.P. Writing ± original draft: P.P. Review & editing: R.H., W.F., S.S., H.N.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pia Palamides, Email: Pia_palamides@web.de.
Rainer Haas, Email: haas@mvp.lmu.de.
References
- 1.Hunt RH, et al. Helicobacter pylori in developing countries. World Gastroenterology Organisation Global Guideline. J Gastrointestin Liver Dis. 2011;20:299–304. [PubMed] [Google Scholar]
- 2.Hooi JKY, et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 3.Weyermann M, Rothenbacher D, Brenner H. Acquisition of Helicobacter pylori infection in early childhood: independent contributions of infected mothers, fathers, and siblings. Am J Gastroenterol. 2009;104:182–189. doi: 10.1038/ajg.2008.61. [DOI] [PubMed] [Google Scholar]
- 4.Bastos J, et al. Sociodemographic determinants of prevalence and incidence of Helicobacter pylori infection in Portuguese adults. Helicobacter. 2013;18:413–422. doi: 10.1111/hel.12061. [DOI] [PubMed] [Google Scholar]
- 5.Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487–490. doi: 10.1002/ijc.28999. [DOI] [PubMed] [Google Scholar]
- 6.Malfertheiner P, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 7.Holcombe C. Helicobacter pylori: the African enigma. Gut. 1992;33:429–431. doi: 10.1136/gut.33.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miwa H, Go MF, Sato N. H. pylori and gastric cancer: the Asian enigma. Am J Gastroenterol. 2002;97:1106–1112. doi: 10.1111/j.1572-0241.2002.05663.x. [DOI] [PubMed] [Google Scholar]
- 9.Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J. Clin. Invest. 2004;113:321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odenbreit S, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 11.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277:6775–6778. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 12.Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003;125:1636–1644. doi: 10.1053/j.gastro.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Basso D, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135:91–99. doi: 10.1053/j.gastro.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 14.Chang WL, Yeh YC, Sheu BS. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25:68. doi: 10.1186/s12929-018-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utsch, C. & Haas, R. VacA’s Induction of VacA-Containing Vacuoles (VCVs) and Their Immunomodulatory Activities on Human T Cells. Toxins. (Basel)8, toxins8060190 [pii], 10.3390/toxins8060190 [doi] (2016). [DOI] [PMC free article] [PubMed]
- 16.McClain, M. S., Beckett, A. C. & Cover, T. L. Helicobacter pylori Vacuolating Toxin and Gastric Cancer. Toxins (Basel)9, 10.3390/toxins9100316 (2017). [DOI] [PMC free article] [PubMed]
- 17.Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 18.Cover TL, Krishna US, Israel DA, Peek RM. Induction of gastric epithelial cell apoptosis by Helicobacter pylori vacuolating cytotoxin. Cancer Res. 2003;63:951–957. [PubMed] [Google Scholar]
- 19.Atherton JC, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J. Biol. Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 20.Sheikh AF, et al. CagA and vacA allelic combination of Helicobacter pylori in gastroduodenal disorders. Microb Pathog. 2018;122:144–150. doi: 10.1016/j.micpath.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Xiang ZY, et al. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infection and Immunity. 1995;63:94–98. doi: 10.1128/IAI.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison, U. et al. Helicobacter pylori strains from a Nigerian cohort show divergent antibiotic resistance rates and a uniform pathogenicity profile. PLoS. ONE12, e0176454, 10.1371/journal.pone.0176454 [doi];PONE-D-16-38434 [pii] (2017). [DOI] [PMC free article] [PubMed]
- 23.Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128:833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes LI, et al. Lack of association between Helicobacter pylori infection with dupA-positive strains and gastroduodenal diseases in Brazilian patients. Int J Med Microbiol. 2008;298:223–230. doi: 10.1016/j.ijmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, et al. The Helicobacter pylori duodenal ulcer promoting gene, dupA in China. BMC Gastroenterol. 2008;8:49. doi: 10.1186/1471-230x-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen LT, et al. Helicobacter pylori dupA gene is not associated with clinical outcomes in the Japanese population. Clin Microbiol Infect. 2010;16:1264–1269. doi: 10.1111/j.1469-0691.2009.03081.x. [DOI] [PubMed] [Google Scholar]
- 27.Abadi, A. T., Taghvaei, T., Wolfram, L. & Kusters, J. G. Infection with Helicobacter pylori strains lacking dupA is associated with an increased risk of gastric ulcer and gastric cancer development. J. Med. Microbiol61, 23–30, jmm.0.027052-0 [pii], 10.1099/jmm.0.027052-0 [doi] (2012). [DOI] [PubMed]
- 28.Delahay RM, Croxall NJ, Stephens AD. Phylogeographic diversity and mosaicism of the Helicobacter pylori tfs integrative and conjugative elements. Mob DNA. 2018;9:5. doi: 10.1186/s13100-018-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer, W. et al. A comprehensive analysis of Helicobacter pylori plasticity zones reveals that they are integrating conjugative elements with intermediate integration specificity. BMC. Genomics15, 310, 1471-2164-15-310 [pii], 10.1186/1471-2164-15-310 [doi] (2014). [DOI] [PMC free article] [PubMed]
- 30.Matsunari O, et al. Rare Helicobacter pylori Virulence Genotypes in Bhutan. Sci Rep. 2016;6:22584. doi: 10.1038/srep22584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 32.Cover TL, Vaughn SG, Cao P, Blaser MJ. Potentiation of Helicobacter pylori vacuolating toxin activity by nicotine and other weak bases. JID. 1992;166:1073–1078. doi: 10.1093/infdis/166.5.1073. [DOI] [PubMed] [Google Scholar]
- 33.Dore MP, Graham DY, Sepulveda AR. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter. 1999;4:154–161. doi: 10.1046/j.1523-5378.1999.99310.x. [DOI] [PubMed] [Google Scholar]
- 34.Gerrits MM, et al. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter. 2006;11:181–187. doi: 10.1111/j.1523-5378.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- 35.Kwon, Y. H. et al. Specific mutations of penicillin-binding protein 1A in 77 clinically acquired amoxicillin-resistant Helicobacter pylori strains in comparison with 77 amoxicillin-susceptible strains. Helicobacter22, 10.1111/hel.12437 (2017). [DOI] [PubMed]
- 36.Rimbara E, Noguchi N, Kawai T, Sasatsu M. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J Antimicrob Chemother. 2008;61:995–998. doi: 10.1093/jac/dkn051. [DOI] [PubMed] [Google Scholar]
- 37.Tseng YS, et al. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur J Clin Invest. 2009;39:807–812. doi: 10.1111/j.1365-2362.2009.02166.x. [DOI] [PubMed] [Google Scholar]
- 38.Korona-Glowniak, I. et al. Antibiotic Resistance and Genotypes of Helicobacter pylori Strains in Patients with Gastroduodenal Disease in Southeast Poland. J Clin Med8, 10.3390/jcm8071071 (2019). [DOI] [PMC free article] [PubMed]
- 39.Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. Diagnosis of Helicobacter pylori: what should be the gold standard? World J Gastroenterol. 2014;20:12847–12859. doi: 10.3748/wjg.v20.i36.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ndububa DA, et al. Correlation between endoscopic suspicion of gastric cancer and histology in Nigerian patients with dyspepsia. Trop. Gastroenterol. 2007;28:69–71. [PubMed] [Google Scholar]
- 41.Fallone CA, Moss SF, Malfertheiner P. Reconciliation of Recent Helicobacter pylori Treatment Guidelines in a Time of Increasing Resistance to Antibiotics. Gastroenterology. 2019;157:44–53. doi: 10.1053/j.gastro.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Laxminarayan R, et al. Access to effective antimicrobials: a worldwide challenge. Lancet. 2016;387:168–175. doi: 10.1016/s0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- 43.Jaka H, et al. The magnitude of antibiotic resistance to Helicobacter pylori in Africa and identified mutations which confer resistance to antibiotics: systematic review and meta-analysis. BMC Infect Dis. 2018;18:193. doi: 10.1186/s12879-018-3099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon DH, et al. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47:2169–2178. doi: 10.1128/aac.47.7.2169-2178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alm RA, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 46.Fischer W, et al. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38:6089–6101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer W, Haas R. The RecA protein of Helicobacter pylori requires a posttranslational modification for full activity. J. Bacteriol. 2004;186:777–784. doi: 10.1128/JB.186.3.777-784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas R, Meyer TF, van Putten JPM. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 51.Schindele, F., Weiss, E., Haas, R. & Fischer, W. Quantitative analysis of CagA type IV secretion by Helicobacter pylori reveals substrate recognition and translocation requirements. Mol. Microbiol100, 188–203, 10.1111/mmi.13309 [doi] (2016). [DOI] [PubMed]
- 52.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 53.Stolte M, Meining A. The updated Sydney system: classification and grading of gastritis as the basis of diagnosis and treatment. Can. J. Gastroenterol. 2001;15:591–598. doi: 10.1155/2001/367832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.