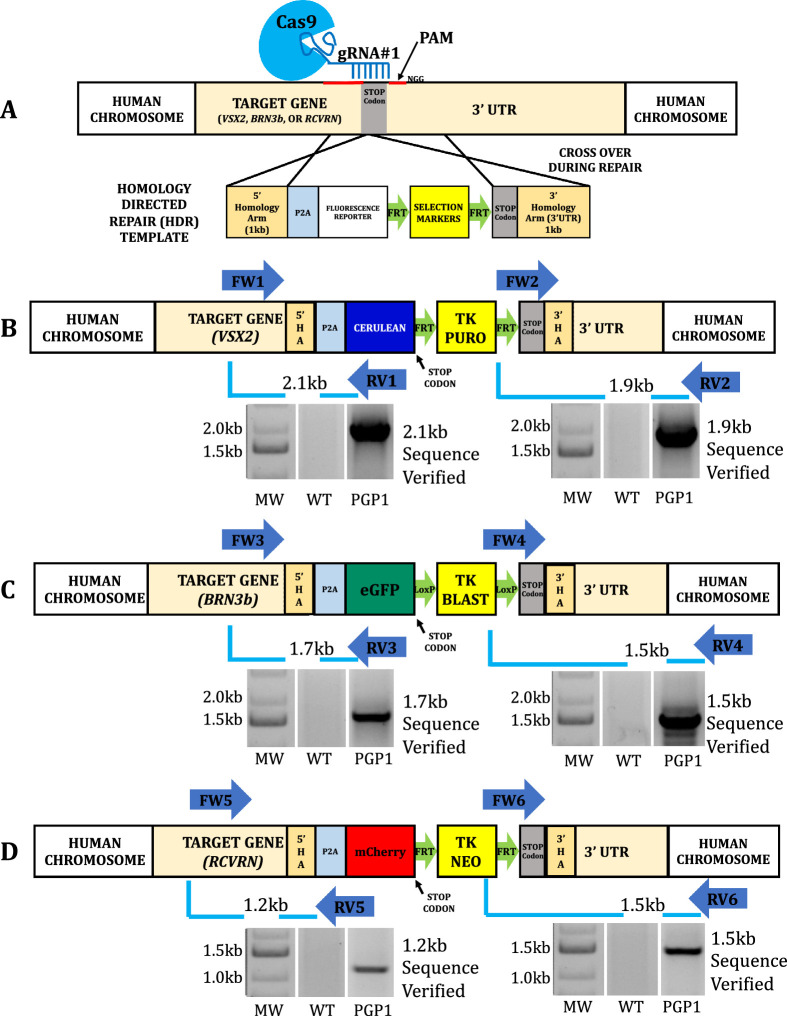
Figure 1.
Creation of the neural retinal progenitor/retinal ganglion cell/photoreceptor (PGP1) reporter hiPSC line by CRISPR/Cas9 genome editing. (A) Schematic illustration of the generalized CRISPR/Cas9-mediated insertion strategy. CRISPR/Cas9 mediated the replacement of the endogenous STOP codon of VSX2 (B), BRN3b (C), and RCVRN (D) loci with P2A:Cerulean, P2A:eGFP, and P2A:mCherry by homologous recombination in WT hiPSCs. Following nucleofection and triple antibiotic selection for puromycin (PURO), blasticidin (BLAST), and G418 (NEO), the resistant clones were screened by PCR with primer sets. (B) FW1/RV1 (forward primer [FW] located outside VSX2 5′HA and reverse primer [RV] located inside Cerulean) with the expected band size of 2.1 kb and FW2/RV2 (inside PURO to outside VSX2 3′HA) with the expected band size of 1.9 kb. (C) FW3/RV3 (outside BRN3b 5′HA to inside membrane tagged eGFP) with the expected band size of 1.7 kb and FW4/RV4 (inside BLAST to outside BRN3b 3′HA) with the expected band size of 1.5 kb. (D) FW5/RV5 (outside RCVRN 5′HA to inside mCherry) with the expected band size of 1.2 kb and FW6/RV6 (inside NEO to outside RCVRN 3′HA) with the expected band size of 1.5 kb. The WT hiPSC was used as control where no bands were seen. All the positive PCR bands were verified by sequencing. The original gels that were cropped for clarity in this figure (with white spaces between nonadjacent lanes) can be seen in their entirety in Supplementary Figure S9. Primer information listed in Supplementary Table S8.