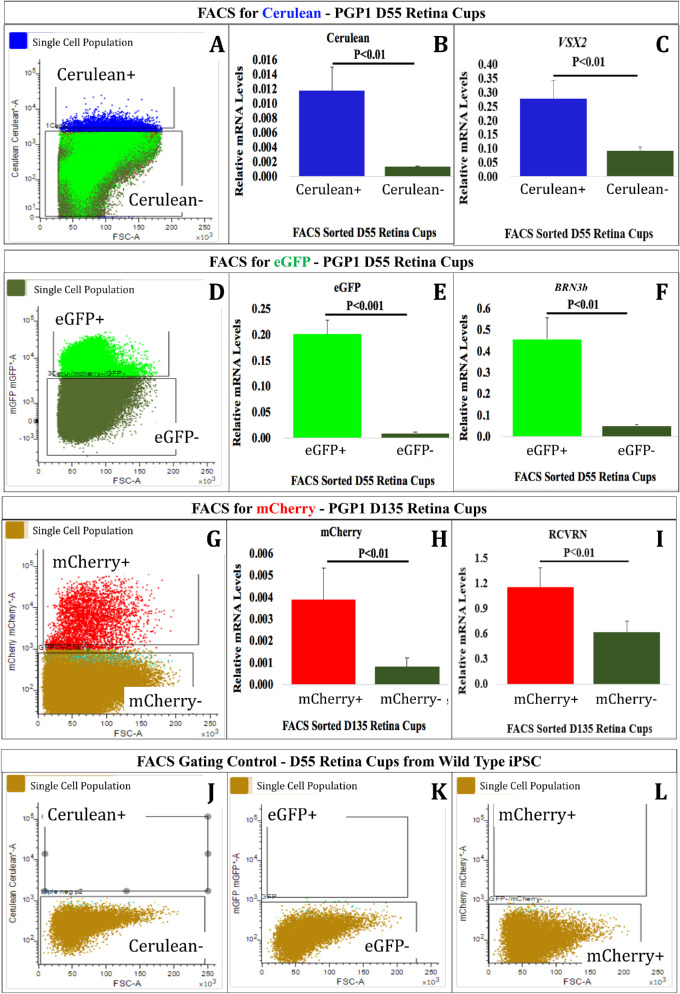
Figure 4.
Confirmation of PGP1-derived retinal organoids corresponded to the appropriately targeted cell types. At D55 (A–F, J–L) and D135 (G–I), the retinal organoids were dissociated into single cells and used for FACS analysis. At D55, (A) the Cerulean+ population was sorted from the Cerulean− population. RT-qPCR analysis revealed that the Cerulean+ cells expressed significantly more Cerulean mRNA (B) and VSX2 mRNA (C) than the Cerulean− cells. Also, at D55, (D) the eGFP+ population was sorted from the eGFP− population. RT-qPCR analysis demonstrated that the eGFP+ cells expressed significantly more eGFP mRNA (E) and BRN3b mRNA (F) than the eGFP− cells. At D135, (G) the mCherry+ population was sorted from the mCherry− population. RT-qPCR analysis revealed that the mCherry+ cells expressed significantly more mCherry mRNA (H) and RCVRN mRNA (I) than mCherry− cells. As a negative control, retinal organoids (D55) derived from WT hiPSC cells were run through the FACS and analyzed with the gates used for Cerulean (J), eGFP (K), and mCherry (L) with no cells occupying those gates. All RT-qPCR data were normalized to GAPDH expression. Error bars represent standard error of the mean (SE). Primer information listed in Supplementary Table S7.