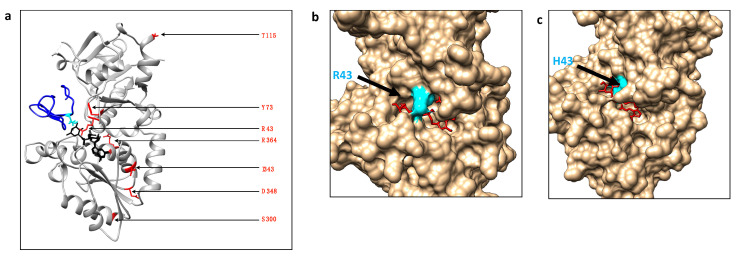
Figure 6.
Location of POFUT1 residues mutated in CRC and automatic docking models for WT POFUT1 and R43H variant with GDP-fucose as a ligand. (a) Using Matchmaker of CHIMERA, X-ray structure of human POFUT1 (grey) co-crystallized with GDP-fucose (black) (PDB 5UXH) was superimposed with mouse POFUT1 co-crystallized with EGF-LD 26 (blue) (PDB 5KY4). After removal of mouse POFUT1, residues implicated in CRC mutations were shown with their sidechains (red). (b) Using the COACH-D docking server, we performed automatic molecular docking using the FASTA sequence of WT human POFUT1 as a template and GDP-fucose (in red) as a ligand (donor substrate). The residue R43, located in front of the binding pocket to GDP-fucose, was colored in cyan. (c) In the same way, a structural model was generated for mutated POFUT1 in which the H43 moved to a more deeply buried position in the binding pocket of GDP-fucose, compared to R43 in WT POFUT1. This could lead to a more exposed fucose moiety of the donor substrate to improve fucose transfer to an EGF-LD.