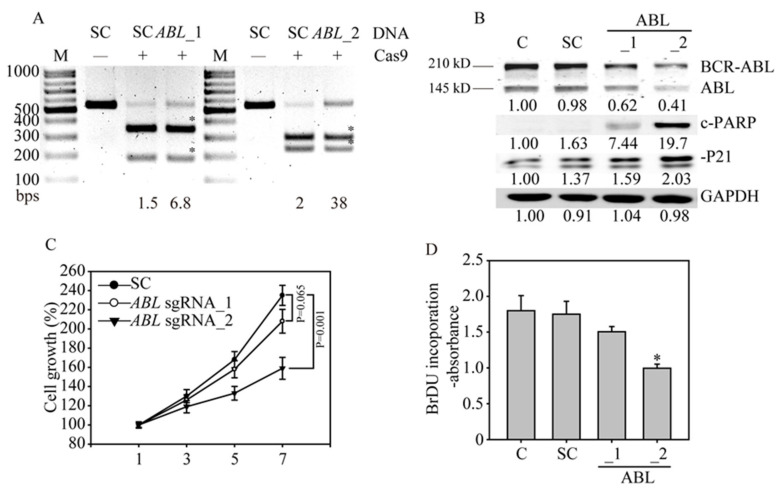
Figure 3.
ABL gene disruption inhibits cancer cell growth and induces apoptosis in K562 cells. (A) The ABL gene in K562 cells was analyzed with RNA-guided engineered nuclease-restriction fragment length polymorphism (RGEN-RFLP) assays to measure the gene editing efficiency. The gel image of ABL gene cleavage induced by addition of specific sgRNA, and Cas9 shows the indel percentage in the gene editing pool. Cleaved DNA fragments are highlighted with an asterisk. (B) Western blot analysis of BCR-ABL and ABL protein expression. Bands at 210 kDa and 145 kDa, corresponding to BCR-ABL and ABL, were observed in the parental and SC sgRNA control K562 cells, whereas these bands were significantly reduced in the ABL sgRNA_1- and ABL sgRNA_2-transduced K562 cells. The protein expression of downstream ABL targets, such as P21 and cleaved PARP (c-PARP), was also activated by ABL gene redundancy. All the western blotting was measured and quantified by Image J software. (C) Cell viability curve of the ABL sgRNA_1- and ABL sgRNA_2-transduced K562 cells determined by MTT assays. (D) Cell proliferation of the ABL sgRNA-transduced K562 cells was determined by bromodeoxyuridine (BrdU) incorporation assays.