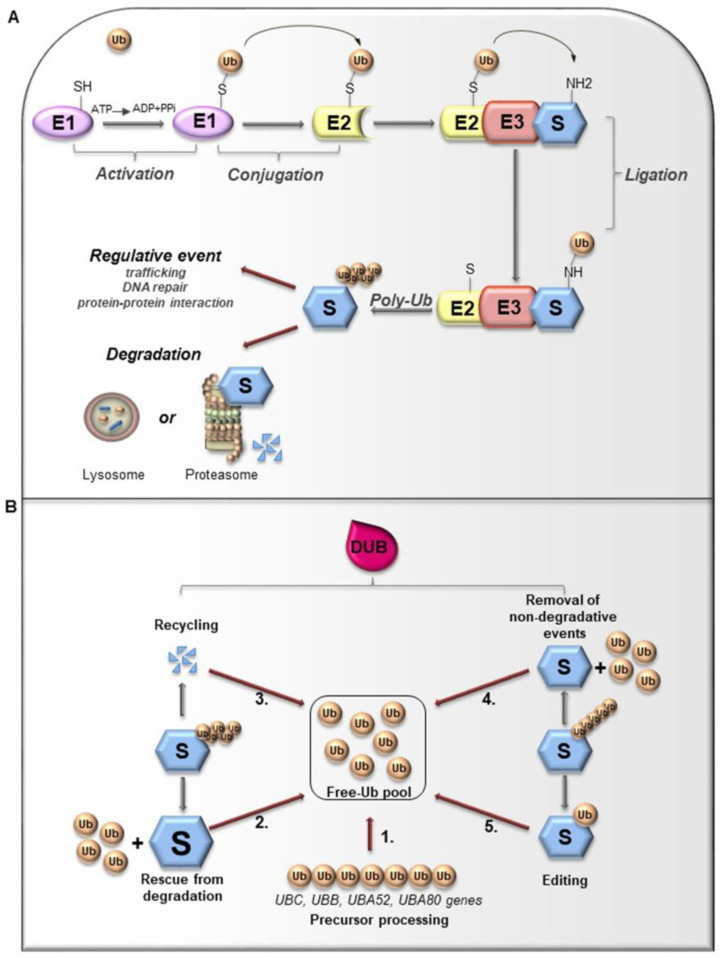
Figure 3.
(A). Ubiquitylation processes. Ubiquitylation is a multi-step process that involves three enzymes: E1 (Ub-activating enzyme), E2 (Ub-conjugating enzyme) and E3 (Ub-ligase). Initially, Ub is linked to E1 through a high energy thioester bond. After, Ub activated by E1 is conjugated to a sulfhydryl group on E2 enzyme. Finally, E3 ligase specifically catalyzes the transfer of Ub from E2 to a Lys residue on a substrate protein. The formation of a poly-ubiquitin (poly-Ub) chain can lead the substrate toward a degradative or regulative pathway. (B). Deubiquitylation and DUBs function. Ubiquitylation can be reversed by deubiquitylating enzymes (DUBs) that hydrolyze the isopeptide or peptide bond, leading to Ub deconjugation from the ubiquitylated protein. DUBs have many functions. 1. Precursor processing: Ub is encoded by four genes and translated as a linear fusion protein consisting of multiple Ub copies, which require the cleavage by DUBs in order to generate free single Ub; 2. Rescue from degradation: DUBs can rescue protein from proteasomal or lysosome degradation; 3. Recycling: DUBs maintain Ub homeostasis preventing its degradation following substrate proteolysis; 4. Removal of non-degradative events: DUBs can remove Ub chains from substrates that are not committed to degradation; 5. Editing: DUBs can also affect the fate of ubiquitylated substrates by cleaving inter-Ub chains (switching from degradative to non-degradative ubiquitylation).