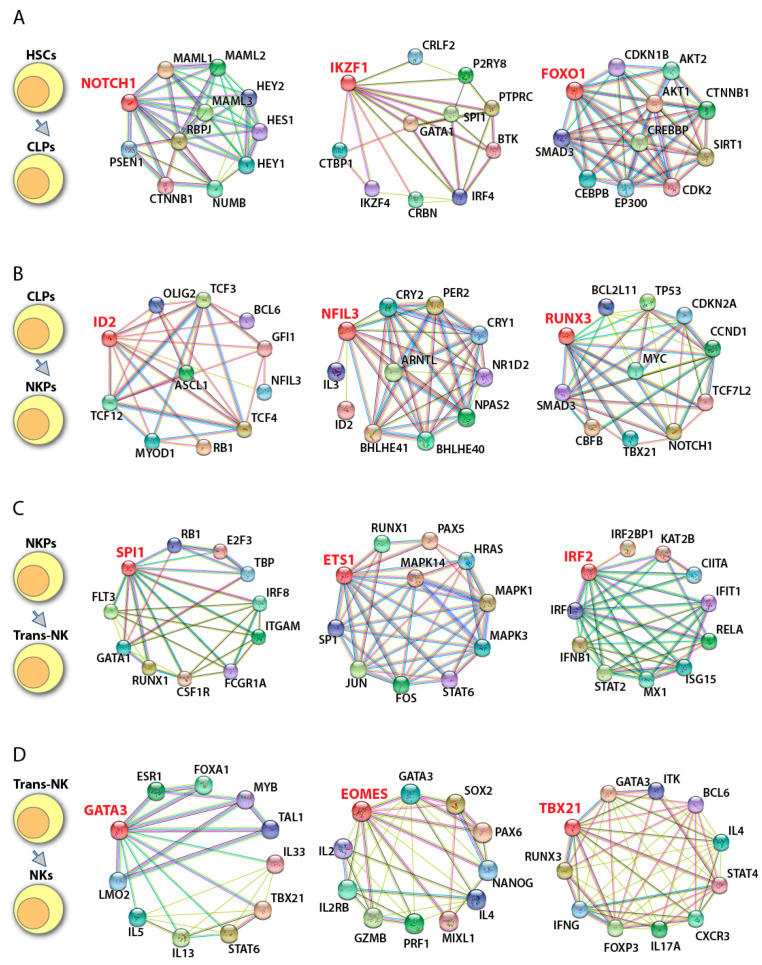
Figure 5.
Potential transcriptional networks in human NK cell development. Potential downstream and upstream regulators of individual transcription factors involved in human NK cell development were identified using String database String database (https://string-db.org/). Gene of interest is shown in red font. ‘—’ represent data from curated databases; ‘—’ experimentally determined; ‘—’ gene neighborhood; ‘—’ gene fusions; ‘—’ gene cooccurrence; ‘—’ text mining; ‘—’ co-expression; and ‘—’ protein homology. (A) Human HSCs transit to CLPs under the help of NOTCH1, IKAROS, and FOXO1. (B) The commitment to NK lineage is mainly regulated by ID2, NFIL3, and RUNX3. (C) Transition from NKPs to iNK is potentially governed by PU.1, ETS-1, and IRF2. (D) The maturation of human NK cells is regulated by GATA3, EOMES, and T-bet.