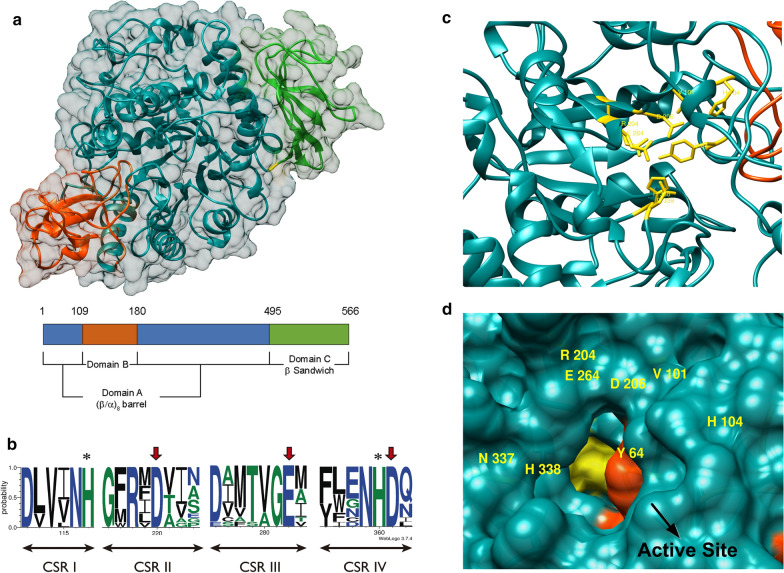
Fig. 2.
Preliminary structural analysis of recombinant α-glucosidases. Superposition of the overall structures of Mg/Mr-αGlu based on the S. cerevisiae oligo-1,6-glucosidase (PDB: 3A47_A) template. The tree typical domains of the glycoside hydrolase family GH13 were depicted using the colour code given in the scheme (a). Sequence logo of the four conserved regions from the family GH13 identified in Mg/Mr-αGlu proteins. Amino acids of the catalytic triad were indicated with red arrows. The rest of highly conserved residues important for the catalytic mechanism were marked with asterisks. The logo was performed using 11 characterised microbial glycoside hydrolases of the family GH13 (PDB: 3A47_A; 1UOK_A; 5DO8_A; 4MB1_A; 5BRQ_A; 5ZCB_A; 2ZE0_A; 4AIE_A; 1M53_A; 1ZJA_A and UniProtKB: Q9P6J3) (b). Schematic view of the conserved residues (shown in yellow) of Mg/Mr-αGlu potentially implicated in subsite -1: Y64, V101, H104, R204, D206, E264, H338 and D339 (c). Surface representation of the active site entrance. Catalytic domain was coloured in dark cyan and domain B in red orange. The subsite -1 surface was highlighted in yellow and residues implicated were labelled (d)