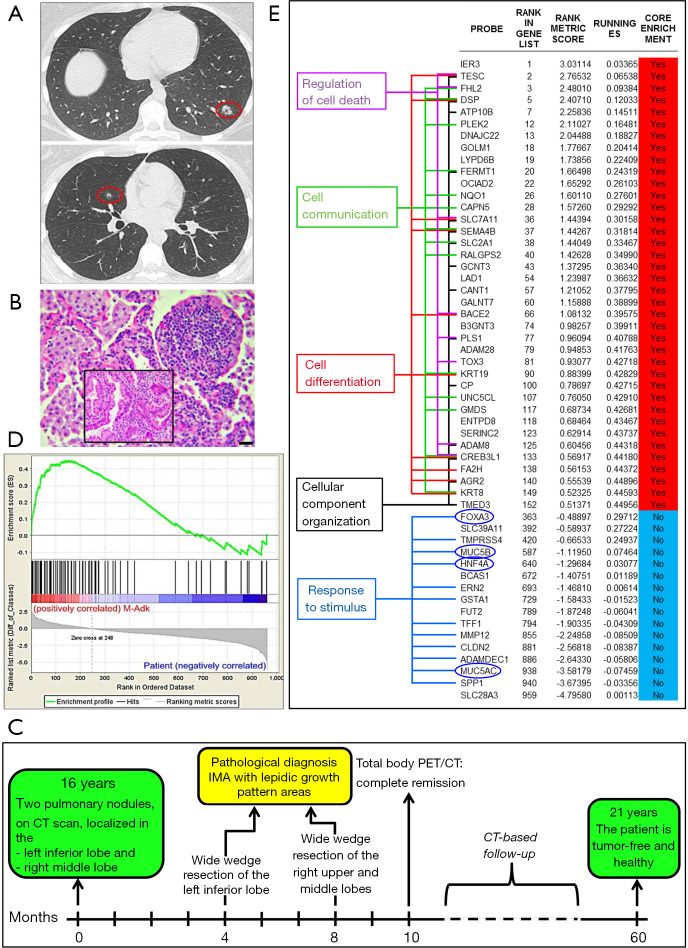
Figure 1.
Radiological, histological and transcriptomic analyses of patient’s IMA. (A) Spiral CT scans demonstrated, in the left lower lobe, a nodular consolidation area, with irregular margins, of 15 mm in size (red circle) and, in the right middle lobe, an excavated nodule of 7 mm in size (red circle). (B) Hematoxylin and eosin staining of one of the lung neoplastic nodules revealed the histologic feature of IMA, with an intratumoral lymphoid follicle-like structure. Magnification: ×200. Scale bar: 30 µm. The inset shows the aspects of lepidic growth and a considerable lymphomononuclear cell infiltrate in the tumor stroma. Magnification: ×400. Scale bar: 40 µm. (C) Timeline of the patient. (D) Gene Set Enrichment Analysis (GSEA) (http://software.broadinstitute.org/gsea/index.jsp) shows that the IMA Signature (143 genes) developed by Guo et al. in 2017, through the analysis of six IMA cases (Enrichment Score =0.4495621; Nominal P value <1%; False Discovery Rate <25%), is not enriched in our patient (Enrichment score <0.1). (E) The table shows the genes, included in the IMA signature, ordered by the normalized enrichment score. Genes with a “Yes value” (in red), contribute to the leading edge subset within the gene set (i.e., genes that contributes most to the enrichment result). Genes with a “No value” (in blue) contribute less to the enrichment result. Genes that regulate cell death, communication, differentiation and cellular component organization prevail in the IMA signature, whereas expression of genes regulating the response to stimulus, including typical IMA-associated genes, such as HNF4A, FOXA3, MUC5AC and MUC5B (circled in dark blue), are prevalent in the patient’s IMA. IMA, invasive mucinous adenocarcinoma.