Figure 6. Four SVs in three MADS-box genes were required to breed for the jointless trait.
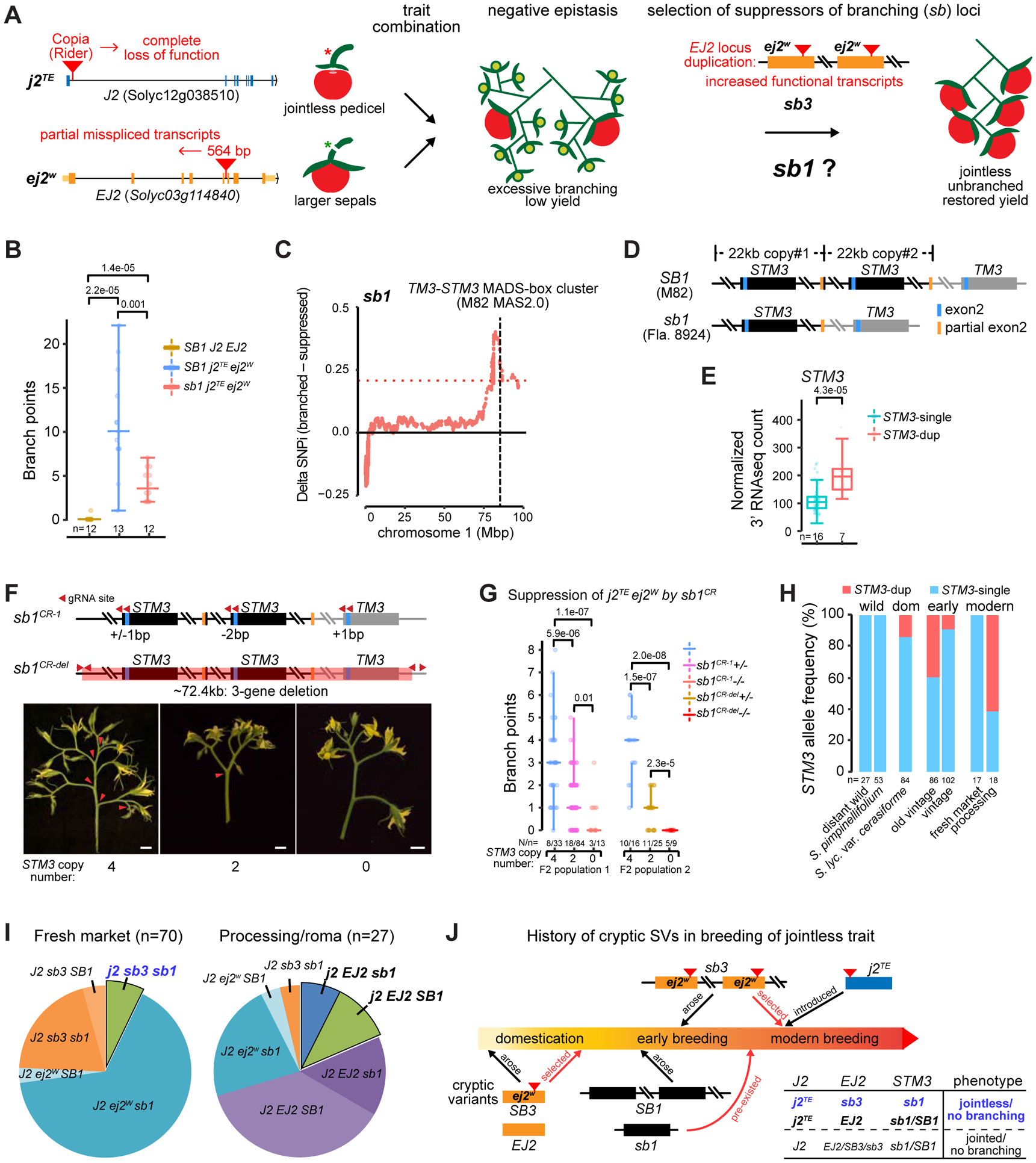
(A) Genetic suppressors were selected to overcome a negative epistatic interaction on yield caused by mutations in two MADS-box genes. The SV mutation j2TE causes a desirable jointless pedicel that facilitates harvesting. Introducing j2TE in backgrounds carrying the cryptic SV mutation ej2w results in excessive inflorescence branching and low fertility. The sb1 and sb3 QTLs were selected to suppress j2TE ej2w negative epistasis. sb3 is an 83 kb duplication harboring ej2w. sb1 is cloned in this study.
(B) Quantification of sb1 partial suppression of branching in the j2TE ej2w background. The SB1 j2TE ej2W and sb1 j2TE ej2W genotypes were derived from F3 families. Each data point is one inflorescence from F4 plants (n).
(C) Delta SNP index (deltaSNPi, QTL-seq) plot shows the sb1 locus contains the TM3-STM3 MADS-box gene cluster (see STAR Methods).
(D) Schematic of the TM3-STM3 locus in the SLL genotypes M82 and Fla.8924, with M82 having a ~22 kb tandem duplication (designated SB1) containing STM3.
(E) RNA-seq showing increased expression of STM3 from the SB1 duplication compared to sb1.
(F) CRISPR-Cas9 mutagenesis of the TM3-STM3 cluster (sb1CR) suppresses branching in the j2TE ej2w background. Schematics at top depict two CRISPR lines with indel mutations in the STM3 and TM3 genes (sb1CR−1) and a large deletion spanning all three genes (sb1CR-del) (top). Representative inflorescences from the indicated genotypes (bottom). Arrowheads mark branch points.
(G) Quantification and comparison of suppression of inflorescence branching by homozygous and heterozygous sb1CR−1 and sb1CR-del mutations in the background of j2TE ej2w. Genotypes were derived from F2 populations (see STAR Methods). N, plant number. n, inflorescence number.
(H) STM3 duplication allele frequency in wild tomato species (distant relatives, SP), early domesticates and cultivars (SLC, SLL vintage) and modern cultivars (SLL fresh market and processing).
(I) Distribution of J2 EJ2 SB1 genotypes in fresh market and processing/roma tomato types. All j2 fresh market genotypes carry sb1 and sb3, whereas processing/roma genotypes have SB1 or sb1, because EJ2 is functional.
(J) Schematic showing the history of breeding for the jointless trait, including when SVs in EJ2 and STM3 arose. The pre-existing sb1 cryptic variant (single copy STM3) mitigated the severity of branching caused by introduction of j2TE in varieties carrying the cryptic variant ej2w. Selection of the sb3 cryptic variant (two copies of ej2w) resulted in the complete suppression of branching and restoration of normal yield. Gradient colored bar represents timeline. The table summarizes genotypic combinations. Blue and black bold fonts indicate solutions for jointless breeding in fresh market and processing/roma types, respectively (I and J).
In (B, E, H and I), n represents sample size. P-values in (B and G) are based on two-tailed, two-sample t-tests. See also Figure S6.
