Graphical abstract
Keywords: Carbon materials, Adhesion, Nanoparticles, Microstructure, Enterovirus
Highlight
-
•
Cellulose microfibers were modified by polyglutamic acid and mesoporous silica nanoparticles.
-
•
The modified microfibers strongly adsorbed the epitope of EV71 capsid.
-
•
The modified microfibers high-efficiently adsorbed EV71 virus particles.
Abstract
In this study, we synthesized a novel kind of cellulose-based microfibers for efficient adsorption of Enterovirus 71 (EV71), the leading causative agent of life-threatening hand, foot and mouth disease. The initial cellulose microfibers (CEL) were activated by (3-aminopropyl) triethoxysilane (APTES), and then covalently modified by polyglutamic acid (PGA) and mesoporous silica nanoparticles (MSN), obtaining the microfibers CEL-PGA-MSN. Owing to the electrostatic interaction between the negatively charged components (i.e., PGA and MSN) and positively charged amino acids of the epitope of EV71 capsid protein VP2 (VP2-ep), the obtained microfibers strongly adsorbed the epitope, and exhibited high EV71-adsorption capacity. This study sheds a novel light on development of cellulose-based materials for application in virus-capturing equipment.
1. Introduction
Viruses transmitted through the gastrointestinal system (e.g., enteroviruses) or the respiratory system (e.g., SARS-Cov-2, MERS, SARS-Cov-1) are becoming world-wide concerns severely threatening human beings, owing to their extremely strong infectivity, high morbidity and high mortality [1], [2]. Among them, Enterovirus 71 (EV71) is a non-enveloped single-strand RNA virus, constituting the dominant causative agent of hand, foot and mouth disease (HFMD) [3]. This serious disease frequently outbreaks in children in the Asia-Pacific region and Europe, and is frequently associated with meningitis, encephalitis, polio-like syndrome leading to high mortality [4]. In 2012, HFMD outbreaks in the mainland China, causing>2 million cases and 567 death [5].
Transmission of EV71 between children is frequently mediated by water, food, and air [6], [7]. Therefore, cleaning of the water and air exposing to children by EV71-adsorption materials may facilitate prevention of EV71 infections. Recently, several EV71-adsorption materials (e.g., graphene oxides, metal ion-binding chitosan) have been developed [8], [9]. However, it remains to be investigated to develop EV71-asorption microfibers for preparation of EV71-capturing membranes and masks.
In this study, we synthesized a readily prepared EV71-adsorbing material based on the natural and low-cost cellulose microfibers (Fig. S1). The initial microfibers were covalently modified by polyglutamic acid (PGA) and mesoporous silica nanoparticles (MSNs), obtaining negatively charged microfibers. The obtained microfibers strongly adsorbed the surface epitope of the EV71 capsid protein VP2, and further exhibited high virus-adsorption activity to EV71 particles. This study enlightens development of cellulose microfiber-based adsorption materials for virus capture.
2. Materials and methods
2.1. Materials
Tetraethyl orthosilicate (TEOS), hexadecyltrimethylammonium bromide (CTAB), (3-aminopropyl) triethoxysilane (APTES), 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide (EDC), and N-hydroxysulfosuccinimide (NHSS) were purchased from Sigma, USA. PGA (50–100 kD) was purchased from Heowns, China. NaOH, 2-morpholinoethanesulfonic Acid (MES) were purchased from Guangfu, China. FITC-tagged VP2-ep peptide (FITC-AGGTGTEDTHPPYK) is synthesized by ChinaPeptides, China. The EV71 virus was isolated from stool samples of HFMD patients under routine surveillance and stored in rhabdomyosarcoma (RD) cells.
2.2. Synthesis and characterization of the modified cellulose microfibers
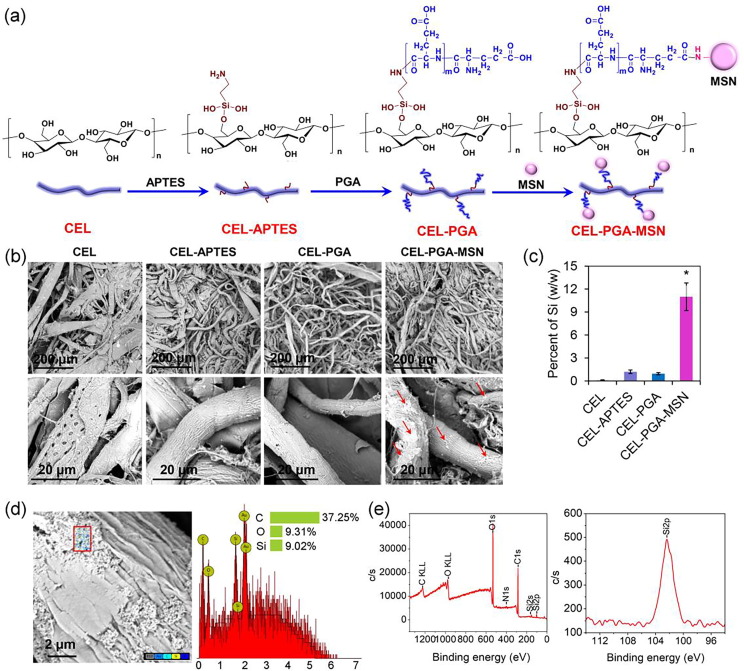
The CEL exposing amino groups was covalently modified by PGA (exposing carboxyl groups) and MSNs (exposing amino groups) by two steps of the EDC/NHSS reaction (Fig. 1 a). Firstly, 1 g of cellulose (CEL) was originally obtained from filter papers, cut and suspended in 100 mL deionized water (dH2O) containing 1.5 g NaOH and 6 g urea. The suspension was vigorously stirred at −10 °C for 12 h, and then the pH was adjusted to pH 4.0 using acetic acid. 5 mL of APTES was dissolved in 25 mL dH2O, and then added into the cellulose suspension. After 24 h of stirring at room temperature, the cellulose microfibers were filtered, washed by dH2O and ethanol, and dried, obtaining CEL-APTES. 1 g of CEL-APTES was suspended in 100 mL of 50 mM MES buffer (pH 6.0), and then 0.2 g of PGA, 0.1 g EDC and 0.11 g NHSS were added into the suspension. The mixture was stirred at room temperature for 24 h, filtered and washed by water, obtaining CEL-PGA. 1 g of CEL-PGA was further suspended in 100 mL of 50 mM MES buffer (pH 6.0), and then 0.15 g of MSNs, 0.1 g EDC and 0.11 g NHSS were added into the suspension. The mixture was stirred for 24 h and washed, obtaining the final CEL-PGA-MSN. The microfibers were characterized by scanning electron microscopy (SEM) with energy dispersive X-ray spectrometry (EDS) mapping (Tecnai G2 F20 S‐TWIN, FEI, USA), inductively-coupled plasma (ICP, Perkin Elmer, USA), Fourier-transform infrared spectroscopy (FT-IR, Vertex 70, Bruker, USA) and XPS analysis (PHI 5000 Versa Probe, ULVAC, Japan).
Fig. 1.
Synthetic route and characterization of the modified cellulose microfibers. (a) Synthetic route of the microfibers. (b) SEM images. The red arrows indicate the MSNs modified onto the microfibers. (c) Silica contents (w/w) of the modified microfibers revealed by ICP assays. * indicates significant difference between the groups (P < 0.05). (d) Amplified SEM image and EDS mapping of CEL-PGA-MSN. (e) XPS spectra of CEL-PGA-MSN. Left, whole spectrum; right, Si2p spectrum. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Adsorption assays of the EV71 epitope
To assess the adsorption ability of the modified microfibers, the solution of the EV71 epitope peptide VP2-ep (in PBS) were added into the microfiber suspension (100 mg/L, prepared in PBS) to a final concentration of 100 mg/L. The mixture was shaken for 24 h and then filtered, obtaining filtering liquid for determining fluorescence intensity by a microplate reader (PerkinElmer, USA).
2.4. Adsorption assays of EV71
100 μL of EV71 solution were diluted in 10 mL dH2O, and then 100 mg of the microfibers were added. The mixture was shaken for 2 h or for indicated time and filtered. The filtered liquid was used for RNA extraction in a nucleotide extractor (QIAcube HT, USA). EV71 nucleotides were detected using the QX100 Droplet Digital PCR (ddPCR) System (Bio-Rad, USA) [11], with the EV71 detecting primers EV71-S (GCAGCCCAAAAGAACTTCAC) and EV71-A (ATTTCAGCAGCTTGGAGTGC).
3. Results
3.1. Characterization of the modified cellulose microfibers
Owing to the presence of positively charged epitopes on the surface of EV71, two negatively charged components, e.g., PGA and MSNs, were covalently grafted to the surface of cellulose microfibers (Fig. 1a). SEM observations revealed that the modified microfibers (i.e., CEL-APTES, CEL-PGA, and CEL-PGA-MSN), as compared to the initial CEL microfibers with a diameter of 30–50 μm, had a decreased diameter (10–20 μm) (Fig. 1b), which was attributed to the dispersion effect of NaOH and urea. Moreover, the final CEL-PGA-MSN showed obvious MSN distribution on the surface of the microfibers. Consistently, ICP assays revealed that CEL-PGA-MSN had much higher Si contents than other microfibers (11% versus < 1.2%) (Fig. 1c). High-resolution SEM and EDS mapping indicated that the MSNs and corresponding Si atoms on the surface of the CEL-PGA-MSN microfibers (Fig. 1d). XPS and FTIR analysis further confirmed the existence of the expected elements (i.e., C, O, N, and Si, Fig. 1e), and the presence of the Si-O and –CO–NH– groups in CEL-PGA-MSN (Fig. S1). These results indicate successful modification of MSNs on the final cellulose microfibers.
3.2. Adsorption of the EV71 epitope by the modified cellulose microfibers
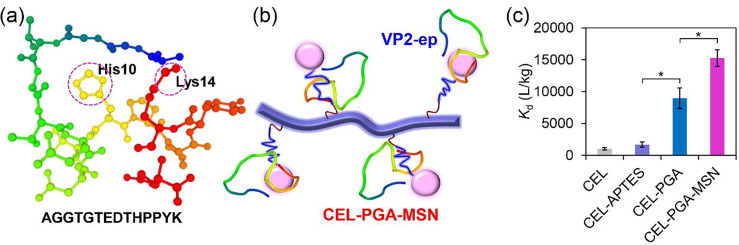
VP2 is one of the most important surface proteins of the EV71 capsid, and the neutralizing epitope VP2-ep (AGGTGTEDTHPPYK) is a representative exposure region [12]. VP2-ep contains two exposed positively charged amino acids (i.e., His10 and Lys14, Fig. 2 a), which may interact with negatively charged PGA and MSNs on CEL-PGA-MSN (Fig. 2b). Epitope adsorption assays were performed to verify the strong interaction between VP2-ep and the negatively charged microfibers. Expectedly, the negatively charged microfibers (i.e., CEL-PGA and CEL-PGA-MSN) had much higher K d values than CEL and CEL-APTES, and CEL-PGA-MSN showed the highest K d value (15255 L/kg) (Fig. 2c). Therefore, the final microfibers had highest activity to bind the EV71 epitope VP2-ep.
Fig. 2.
Adsorption of VP2-ep by the modified cellulose microfibers. (a) Structure of VP2-ep. The dotted purple circles indicate the positively charged groups in the epitope. (b) Adsorption model of VP2-ep binding to CEL-PGA-MSN, indicating the interaction between the VP2-ep peptide and the microfibers through the positively charged amino acids and the negatively charged PGA/MSNs. (c) Kd values of VP2-ep to microfibers. Kd is the distribution coefficient of peptides between adsorbed and solution phases (L/kg), calculated as (C0 − Ce)/(Cmicrofiber·Ce), where C0 is the initial VP2-ep concentration in solution, Ce is its equilibrium concentration in solution, and Cmicrofiber is the microfiber concentration in solution. * indicates significant difference between the groups (P < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Adsorption of EV71 by the modified cellulose microfibers
The high binding activity of CEL-PGA-MSN to the EV71 VP2 epitope suggests its high EV71-adsorption ability. To verify this, the virus was firstly cultured in RD cells, which displayed obvious shrinking morphology as compared to un-infected RD cells (Fig. 3 a), and then isolated from the host cells for adsorption assays. ddPCR analysis revealed that as compared to the supernatant samples after CEL adsorption that exhibited highest positive droplets (blue colors), the supernatants after adsorption by the modified microfibers had obvious decreased positive droplets, and the supernatants after CEL-PGA-MSN adsorption had the least positive droplets (Fig. 3b). Statistical analysis further showed that CEL-PGA-MSN adsorbed highest levels of EV71 (>80%) among these microfibers after 2 h (Fig. 3c). Time-dependent experiment further showed that the adsorption capacity increased with increase in incubation time, and reached > 95% after 4 h (Fig. S2). Moreover, even after 5 operating times, the microfibers kept the adsorption capacity > 65% (Fig. S3), indicating its good reusability. Therefore, CEL-PGA-MSN had the strongest ability to adsorb EV71 virus. [10]
Fig. 3.
Adsorption of EV71 by the modified cellulose microfibers. (a) Microscopic images of the RD cells and EV71-infected RD cells. (b) Droplet digital PCR (ddPCR) analysis of the contents of EV71 nucleotides in the EV71 solution after microfiber adsorption. (c) Statistical analysis of adsorbed EV71 by the microfibers. * indicates significant difference between the groups (P < 0.05).
In conclusion, we prepared a novel kind of cellulose microfibers by covalent co-modification of negatively charged polymers (PGA) and inorganic nanoparticles (MSN). The obtained CEL-PGA-MSN showed excellent adsorption capacity to the EV71 VP epitopes and consequent EV71 virus. This study sheds a novel light on development of low-cost, reusable, and high-efficient virus-removing materials.
CRediT authorship contribution statement
Meiqing Sun: Conceptualization, Investigation, Supervision, Methodology, Software, Writing - original draft, Writing - review & editing. Hong Wang: Data curation, Investigation, Writing - review & editing. Xiaoyan Li: Software, Validation, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work is supported by Project of Science and Technology Development in Wuqing District, Tianjin (WQKJ201972).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.matlet.2020.128320.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Carroll D., Daszak P., Wolfe N.D., Gao G.F., Morel C.M., Morzaria S., Mazet J.A. Science. 2018;359(6378):872. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- 2.de Crom S.C., Rossen J.W.A., Van Furth A.M., Obihara C.C. Eur. J. Pediatr. 2016;175(8):1023. doi: 10.1007/s00431-016-2725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Guo Q. New Engl. J Med. 2020;382(12):1177. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang K.Y.A., Chen M.F., Huang Y.C., Shih S.R., Chiu C.H., Lin J.J., Lin T.Y. Nat. Commun. 2017;8(1):762. doi: 10.1038/s41467-017-00736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Zou G., Xia A., Wang X., Cai J., Gao Q., Altmeyer R. Virol. J. 2015;12(1):83. doi: 10.1186/s12985-015-0308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran C.B.N., Nguyen H.T., Phan H.T.T., Van Tran N., Wills B., Farrar J., Simmons C.P. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0021116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W.C., Shih W.L., Yang S.C., Yen T.Y., Lee J.T., Huang Y.C., Huang L.M. J. Microbiol. Immun. Infect. 2017;50(1):10. doi: 10.1016/j.jmii.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y.C., Lin S.T., Chen C.Y., Wu S.C. Enterovirus 71 adsorption on metal ion-composite chitosan beads. Biotechnol. Prog. 2012;28(1):206. doi: 10.1002/btpr.699. [DOI] [PubMed] [Google Scholar]
- 9.Song Z., Wang X., Zhu G., Nian Q., Zhou H., Yang D., Tang R. Small. 2015;11(9–10):1171–1176. doi: 10.1002/smll.201401706. [DOI] [PubMed] [Google Scholar]
- 10.Ferris D.P., Lu J., Gothard C., Yanes R., Thomas C.R., Zink J.I. Small. 2011;7(13):1816. doi: 10.1002/smll.201002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whale A.S., Devonshire A.S., Karlin-Neumann G., Regan J., Javier L., Cowen S., Berger A.W. Anal. Chem. 2017;89(3):1724. doi: 10.1021/acs.analchem.6b03980. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Peng W., Ren J., Hu Z., Xu J., Lou Z., Walter T.S., Rao Z. Nat. Struct. Mol. Biol. 2012;19(4):424. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.