Abstract
Cutaneous T cell lymphoma (CTCL) is a rare but potentially devastating primary cutaneous lymphoma. CTCL is characterized by localization of neoplastic T lymphocytes to the skin, with mycosis fungoides (MF) and its leukemic form, Sézary syndrome (SS) being the most common variants. Thymocyte selection-associated high-mobility group box (TOX) gene has been found to be highly expressed in MF and SS. It is reported that higher expression levels of TOX in patients will increase risks of disease progression and poor prognosis. However, the molecular events leading to these abnormalities have not been well understood. To better understand the molecular mechanism underlying TOX-mediated differentially expressed genes (DEGs) in CTCL, and to identify DEGs pathways triggered after knockdown of TOX gene in the CTCL cell line Hut78, we employed two shRNA-mediated lentiviruses to knock down TOX gene in the skin lymphoma cell line HuT78. RNA sequencing (RNAseq) analysis was applied to analyze DEGs, DEGs GO and their corresponding pathways. Knockdown of TOX can induce upregulation of 547 genes and downregulation of 649 genes, respectively. HOXC9 was the most significant downregulated gene. Most DEGs are enriched in malignancies and relate to the Wnt and mTOR signaling pathways, and therefore they can regulate cellular processes and induce different biological regulation. Transcriptome analysis of DEGs after knockdown of TOX in our study provides insights into the mechanism of TOX in CTCL and suggests candidate targets for therapy of CTCL.
Electronic supplementary material
The online version of this article (10.1007/s00403-019-02000-0) contains supplementary material, which is available to authorized users.
Keywords: Cutaneous T cell lymphoma, TOX, RNA sequencing analysis, Differentially expressed gene, Signaling pathway
Introduction
CTCLs are a heterogeneous group of non-Hodgkin lymphoproliferative disorders characterized by accumulation and expansion of neoplastic T lymphocytes to the skin [28]. MF and SS constitute two main subtypes of CTCL. While MF primarily affects the skin, SS is characterized by the presence of circulating malignant Sézary cells. Together, MF and SS account for 65–80% of CTCL cases [9, 11, 23]. Although accumulative evidence indicates that defects in apoptosis and cell cycle control are critical in disease pathogenesis [5, 21], the molecular mechanism leading to these abnormalities has not been well understood yet.
The TOX gene was firstly described in 2002 [27], as a part of the superfamily of high-mobility group box proteins that act as regulators of gene expression, mainly by modifying the chromatin structure [10, 27]. TOX mRNA is most abundant in the thymus, liver and brain [27]. TOX is involved in lymphocyte maturation, and Zhang et al. demonstrated that TOX was highly and specially expressed in early MF [31]. After this, several studies have confirmed that TOX is aberrantly expressed in CD4+ /CD8− neoplastic T cells in MF and SS [2, 6, 7, 15, 19, 20, 29], so as to be aberrantly expressed in CTCL with CD4−/CD8+ and CD4−/CD8− phenotypes [24], differentiating malignant from non-malignant skin-infiltrating T cells found in benign inflammatory dermatoses [31]. Aberrant expression of TOX plays a central role in malignant survival, proliferation, and tumor formation in CTCL [7]. Stable knockdown of TOX in CTCL cells has promoted apoptosis and reduced cell cycle progression, leading to less cell viability and colony-forming ability in vitro and reducing tumor growth in vivo [7].
It is generally believed that abnormal gene expression is a key process in disease initiation and progression. Hut78 cell line derived from SS exhibits high expression of TOX, and TOX-deficient Hut78 cells can promote apoptosis and reduce cell cycle [7], but its mechanism is not very clear. Herein, we applied RNAseq analysis to further explore transcriptional changes including expressed genes (DEGs), DEG Gene Ontology (GO) and pathways in TOX-deficient Hut78 cells.
Material and methods
Cell culture
Human CTCL cell line Hut78 (ATCC no. TIB161) was cultured in RPMI 1640 and 10%FBS as described by the American Type Culture Collection (Manassas, VA). Infected CTCL cells were cultured in the above medium plus puromycin.
Lentivirus infection
Lentivirus vector (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) and shRNA sequence were designed and synthesized by Genechem (Shanghai, China). Destination cells were infected with lentiviral supernatants, using 8 mg/ml polybrene and high virus titer for MOI ≥ 100. After 48–72 h of incubation, the supernatant was replaced by a medium containing 1 mg/ml puromycin.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cell pellets using TRIzol (Invitrogen, Thermo Fisher) according to the manufacturer’s protocol. cDNA synthesis was performed using the GoScript™ Reverse Transcription System Kit (A5000) from Promega. qPCR reactions were performed with FastStart Universal SYBR Green Master (Rox) from Roche. The experiments were performed according to the manufacturer’s instructions. The sequences of the primers used for qRT-PCR analyses are listed in Table S3. All reactions were run in triplicate. The CT values were calculated using the standard curve method.
Western blotting
After lentivirus infection, HuT78 cell pellets were prepared by centrifugation at 300g, and then total cells were lysed in RIPA buffer (50 mM Tris, 150 mM sodium chloride, 1% Triton X-100, 0.1% sodium dodecyl sulfate, and 1% sodium deoxycholate). After removing insoluble material by centrifugation at 10,000g at 4 ℃ for 5 min, total protein concentration was determined using BCA assay as per manufacturer’s instruction with a microplate reader. 40 μg protein was used for SDS-PAGE gel electrophoresis (Bio-Rad) and transferred onto PVDF membranes (Bio-Rad). Blocking was done with 5% milk and then the membranes were incubated with primary antibodies, anti-TOX (1:1000 HPA018322, Sigma-Aldrich) or anti-actin (1:5000, A1978, Sigma-Aldrich) overnight at 4 ℃. After washing, membranes were incubated with secondary antibodies (peroxidase-conjugated, suitable for each primary antibody) for 2 h at room temperature. The signal was detected using Bio-Rad ChemiDoc XRS + System after adding Super Signal West Pico chemiluminescence.
Apoptosis detection
The treated Hut78 cells (1 × 106) using shRNA 1 construct were transferred to a 15 ml centrifuge tube. Annexin V binding buffer was added. After centrifugation at 2000 rpm for 5 min at 4 °C, the cells were washed three times and 100 µl of binding buffer, 5 µl of Annexin V-APC and 10 µl of 7-AAD stain (Thermo Fisher Scientific, Inc) were added and incubated in the dark for 25 min. Detection of apoptotic cells was performed by flow cytometry.
Cell cycle analysis
The treated Hut78 cells (1 × 106) using shRNA 1 construct were collected and fixed with 75% ice-cold ethanol at 4 °C overnight and then stained with 5 µl propidium iodide (Thermo Fisher Scientific, Inc.) at room temperature for 5 min in the dark. The cell cycle distribution was analyzed by flow cytometry.
RNAseq analysis
Total RNA from infected cells was harvested and extracted by using TRIzol (Invitrogen, Thermo Fisher). Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit) was used to perform quality control of the total RNA samples:RNA concentration, RIN value, 28S/18S and the fragment length distribution. mRNAs were isolated from total RNA with the oligo(dT) method. The mRNAs were fragmented and then first-strand cDNA and second-strand cDNA were synthesized. cDNA fragments were purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. cDNA fragments were next linked with adapters. Those cDNA fragments with suitable size were selected for the PCR amplification. Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System were used in quantification and qualification of those libraries. Equimolar pooling of libraries was performed based on qPCR values and loaded onto an Illumina Hiseq platform (BGI, China).
Results
Genetic silencing of Tox in Hut78 cells
To investigate the transcriptional changes after TOX knockdown, two lentivirus targets were designed to knock down TOX gene in Hut78 cell line, as presented in Table S2. After lentivirus infection, RT-qPCR and Western blot were completed. TOX expression was significantly reduced in mRNA level as shown in Fig. 1a: Compared to the NC group, both sh1 and sh2 groups demonstrate significantly reduced TOX mRNA expression (p < 0.05). TOX protein expression was also diminished as shown in Fig. 1b with the sh1 group showing more inhibition of TOX expression than the sh2 group. Annexin V-APC/7AAD flow cytometry assay was employed to analyze cell apoptosis, and we observed that apoptotic cells were increased after knockdown of TOX as shown in Fig. 1c. The cell cycle distribution analysis showed more cells in G0/G1 phase and less cells in G2/M phase after knockdown of TOX as shown in Fig. 1d.
Fig. 1.
Lentivirus infection knockdown TOX gene expression. TOX knockdown by 2 shRNAs (sh1 and sh2, both specific for TOX mRNA) and negative control (a non-targeting shRNA). Infected cells were selected by puromycin (1 mg/mL) for 5 days. mRNA and protein were extracted for further analysis. a RT-qPCR was performed between group NC, sh1 and sh2, and primer qGAPDH and TOX were used. TOX was significantly reduced in sh1 (p value = 0.0114, R2 = 0.8305) and sh2 (p value = 0.0286, R2 = 0.7371). b Western blotting was performed with antibodies against TOX and actin proteins. *p < 0.05 by two-tailed Student’s t test with Welch correction. Error bars indicate standard error of the mean. Data shown here are representative of at least three independent experiments. c Annexin V-APC/7AAD flow cytometry assay showed that apoptotic cells were increased after knockdown of TOX. d Annexin/PI flow cytometry assay showed that more cells in the G0/G1 phase and less cells in the G2/M phase after knockdown of TOX
DEGs after TOX knockdown
After RNAseq and reads filtering, we mapped clean reads to reference genome by using Bowtie 2 [12] and then calculated the gene expression level for each sample with RSEM [13], a software package for estimating gene and isoform expression levels from RNAseq data. Subsequently, we calculated Pearson correlation between all samples by using cor, performed hierarchical clustering between all samples by using hclust, performed PCA analysis with all samples using princomp, and drew the diagrams with ggplot2 with functions of R. The number of genes and transcripts in each sample are shown in Table1. We further calculated the heat map of Pearson correlation among all samples, shown in Fig. S1a. Based on the expression information, we performed box plot analysis to show the distribution of the gene expression level of each sample, so that we could observe the dispersion of the distribution (results as shown in Fig. S1b). Based on the gene expression level, we could identify the DEGs between samples or groups. MA plots were used to show the distributions of DEGs in Fig. S1. Compared to the NC group, 3897 genes were overexpressed and 2702 genes were underexpressed in group sh1 (Fig. S1c). Compared to the NC group, 2723 genes were overexpressed and 3224 genes were underexpressed in group sh2 (Fig. S1d). Taken together, after TOX knockdown, a total of 547 genes were upregulated and 649 genes were downregulated. The top 20 downregulated genes are listed in Table 2 and the top 20 upregulated genes are listed in Table S1. Interestingly, we found that multiple genes in the HOX gene family were downregulated in TOX-deficient Hut78 cells.
Table 1.
Genes and transcripts statistics
| Sample | Total gene number | Known gene number | Novel gene number | Total transcript number | Known transcript number | Novel transcript number |
|---|---|---|---|---|---|---|
| NC_1 | 16,143 | 13,262 | 2881 | 28,211 | 14,795 | 13,416 |
| NC_2 | 15,472 | 12,807 | 2665 | 25,537 | 13,585 | 11,952 |
| NC_3 | 15,468 | 12,863 | 2605 | 25,801 | 13,699 | 12,102 |
| sh1_1 | 15,089 | 12,530 | 2559 | 24,071 | 12,629 | 11,442 |
| sh1_2 | 14,639 | 12,088 | 2551 | 21,614 | 11,028 | 10,586 |
| sh1_3 | 15,023 | 12,387 | 2636 | 23,159 | 11,918 | 11,241 |
| sh2_1 | 16,040 | 13,100 | 2940 | 28,513 | 15,031 | 13,482 |
| sh2_2 | 16,281 | 13,395 | 2886 | 30,287 | 16,217 | 14,070 |
| sh2_3 | 13,278 | 11,548 | 1730 | 20,495 | 11,131 | 9364 |
Sample sample name, total gene number the amount of all genes, known gene number the amounts of known genes, novel gene number the amounts of novel genes, total transcript number the amount of all transcripts, known transcript number the amounts of known transcripts, novel transcript number the amounts of novel transcripts
Table 2.
Top 20 significantly downregulated genes after TOX knockdown
| Gene name | GeneID | Lengtha | NC-expressionb | sh1-expression | log2 Ratio (sh1/NC)c | q valued | p valuee | sh2-expression | log2 Ratio (sh2/NC) | q value | p value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HOXC9 | 3225 | 1528 | 90,574 | 16,021 | − 1.837919802 | 0 | 0 | 16,747 | − 2.788427035 | 0 | 0 |
| PIGS | 94005 | 2643 | 54,217 | 7133 | − 2.264951439 | 0 | 0 | 9093 | − 2.929149771 | 0 | 0 |
| TXNDC16 | 57544 | 2854 | 3017 | 68 | − 4.810223171 | 0 | 0 | 313 | − 3.622112876 | 0 | 0 |
| GPATCH4 | 54865 | 2158 | 8047 | 2539 | − 1.002977669 | 6.4747E-244 | 7.0734E-246 | 4285 | − 1.26238857 | 0 | 0 |
| CKB | 1152 | 1475 | 3529 | 984 | − 1.18131624 | 6.1571E-140 | 1.7733E-141 | 907 | − 2.313317716 | 0 | 0 |
| UNC119B | 84747 | 4423 | 3865 | 1210 | − 1.014248397 | 7.3431E-120 | 2.5525E-121 | 2313 | − 1.09393589 | 4.2E-189 | 2.3E-190 |
| ANP32E | 81611 | 3446 | 1085.54 | 125.58 | − 2.450521296 | 1.8879E-119 | 6.6251E-121 | 207.8 | − 2.73837807 | 3.3E-188 | 1.8E-189 |
| GBA | 2629 | 2637 | 795.64 | 60 | − 3.067868433 | 7.1629E-111 | 2.7981E-112 | 422.83 | − 1.26527081 | 3.31E-50 | 8.35E-51 |
| HOXC5 | 3222 | 1645 | 592 | 1 | − 8.548240396 | 5.00096E-96 | 2.45025E-97 | 18 | − 5.392761106 | 7.7E-159 | 4.9E-160 |
| HOXC4 | 3221 | 1689 | 1196 | 281 | − 1.428362385 | 2.4963E-64 | 2.18996E-65 | 323 | − 2.241844062 | 3E-165 | 1.9E-166 |
| PRR5-ARHGAP8 | 553158 | 1179 | 342.35 | 11.31 | − 4.258588209 | 1.93483E-61 | 1.81269E-62 | 97.19 | − 2.169824969 | 1.43E-46 | 4.03E-47 |
| POR | 5447 | 2509 | 860.3 | 170.31 | − 1.675463721 | 1.45546E-58 | 1.43585E-59 | 216.96 | − 2.340643496 | 1.6E-125 | 1.3E-126 |
| MTPN | 136319 | 3900 | 314.21 | 0 | − 8.634372317 | 3.90708E-51 | 4.87628E-52 | 0 | − 9.648818028 | 1.19E-62 | 2.22E-63 |
| PMF1 | 11243 | 1122 | 1023.19 | 273.66 | − 1.241404619 | 7.84823E-45 | 1.21854E-45 | 641.82 | − 1.02606616 | 2.8E-46 | 7.97E-47 |
| CDRT4 | 284040 | 2515 | 243.61 | 0 | − 8.267216577 | 1.33826E-41 | 2.39238E-42 | 0 | − 9.281662288 | 2.29E-51 | 5.62E-52 |
| SDSL | 113675 | 1311 | 325 | 36 | − 2.513157937 | 7.66449E-38 | 1.61629E-38 | 40 | − 3.375600555 | 1.42E-69 | 2.35E-70 |
| CDKN2A | 1029 | 1218 | 1132 | 351 | − 1.028118053 | 6.07066E-37 | 1.30831E-37 | 191 | − 2.920462157 | 2E-209 | 9.4E-211 |
| PCSK1N | 27344 | 1071 | 855 | 238 | − 1.183749877 | 3.87281E-35 | 9.09008E-36 | 178 | − 2.617279921 | 1.5E-141 | 1.1E-142 |
| SURF1 | 6834 | 1046 | 189.76 | 0 | − 7.906819136 | 6.41566E-34 | 1.60992E-34 | 0 | − 8.921264847 | 3.28E-42 | 1.05E-42 |
| ZNF616 | 90317 | 4386 | 295.55 | 35.94 | − 2.378526937 | 1.48496E-32 | 4.00162E-33 | 18 | − 4.390566151 | 3.72E-75 | 5.49E-76 |
aGene length; bgene expression of group NC; clog2 transformed fold change between NC group and sh1 group; dadjusted p value; ep value
GO analysis of DEGs
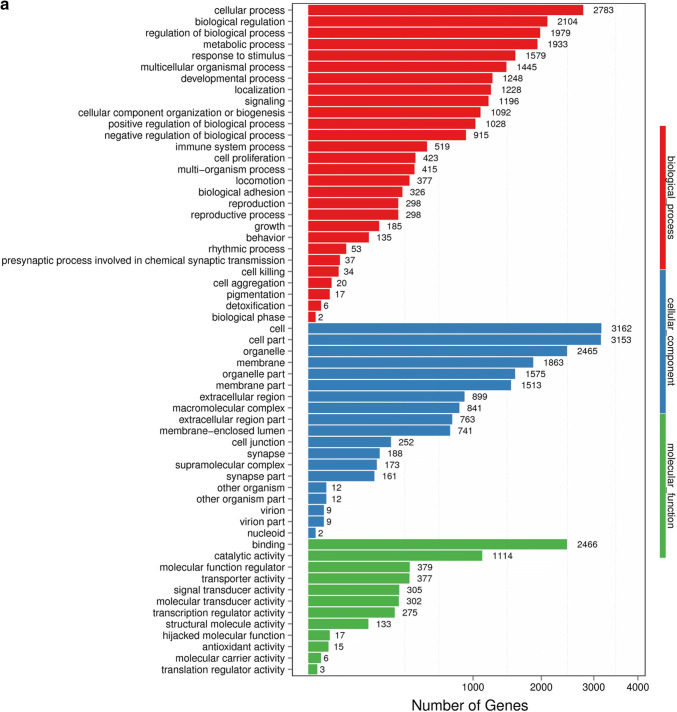
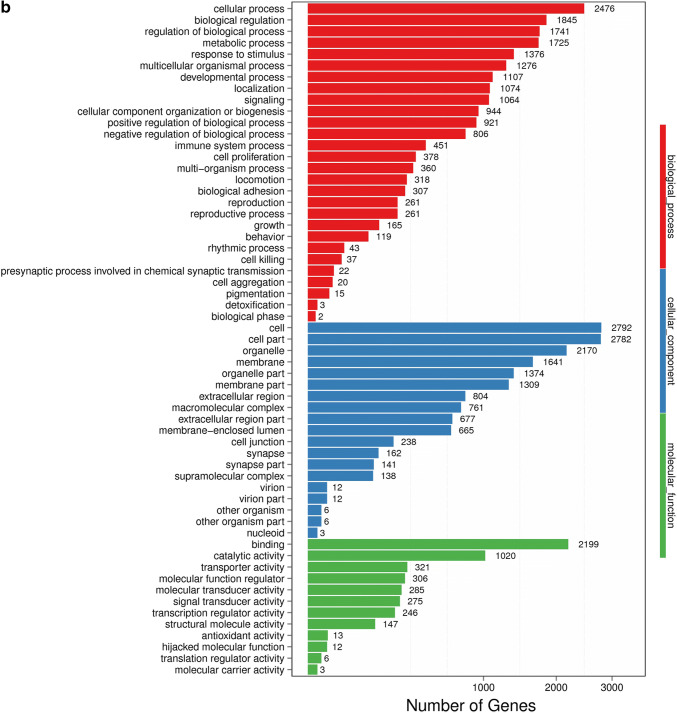
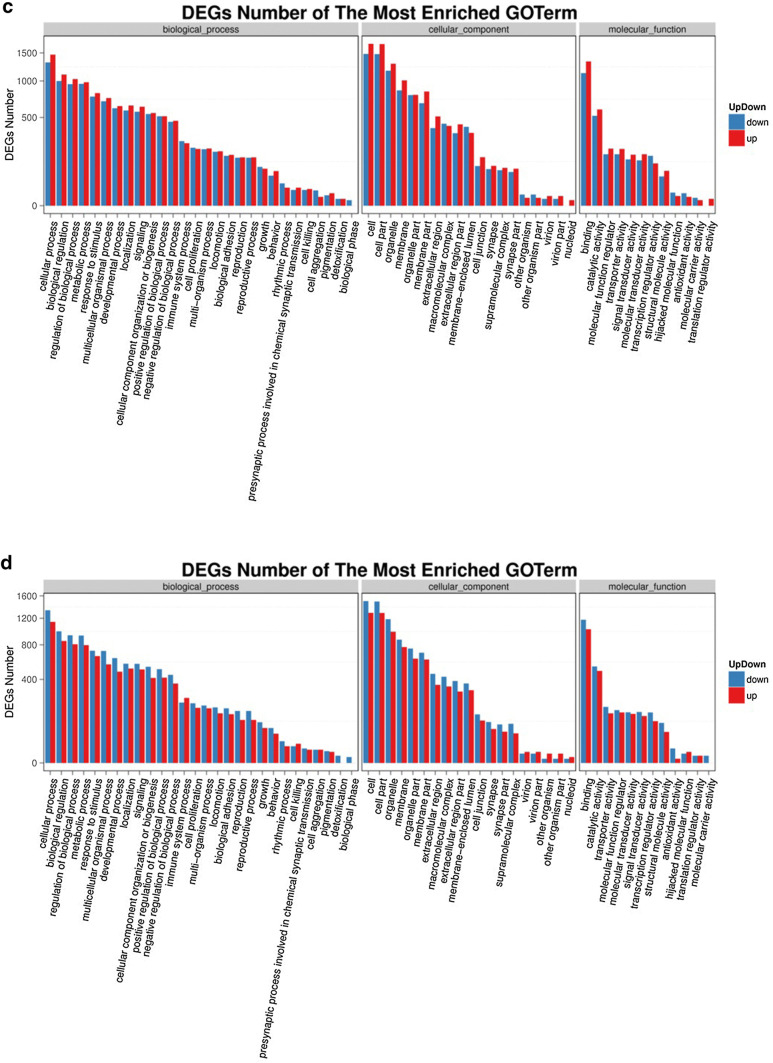
With DEGs, we performed Gene Ontology (GO) classification and functional enrichment. GO has three main ontologies: molecular biological function, cellular component and biological process. The GO classification results are shown in Fig. 2a, b. We used DAG (directed acyclic graph) to show the GO enrichment result. Each bar shows GO terms, and the amount of up- or down-regulated genes are shown in Fig. 2c, d. In our study, we found that TOX gene knockdown could significantly influence the cellular process, the cell growth as well as the death signal transduction, as was previously reported [7]. Among most of the enriched GO terms, most DEGs were related to cellular process, biological regulation and binding process.
Fig. 2.
GO classification of DEGs. a NC vs sh1; b NC vs sh2. GO classification and functional enrichment among molecular biological functions, cellular components and biological processes. X axis represents the number of DEGs. Y axis represents GO terms. c NC vs sh1; d NC vs sh2. GO classification of upregulated and downregulated genes. X axis represents GO terms. Y axis represents the amount of up- or down-regulated genes
Pathway analysis of DEGs
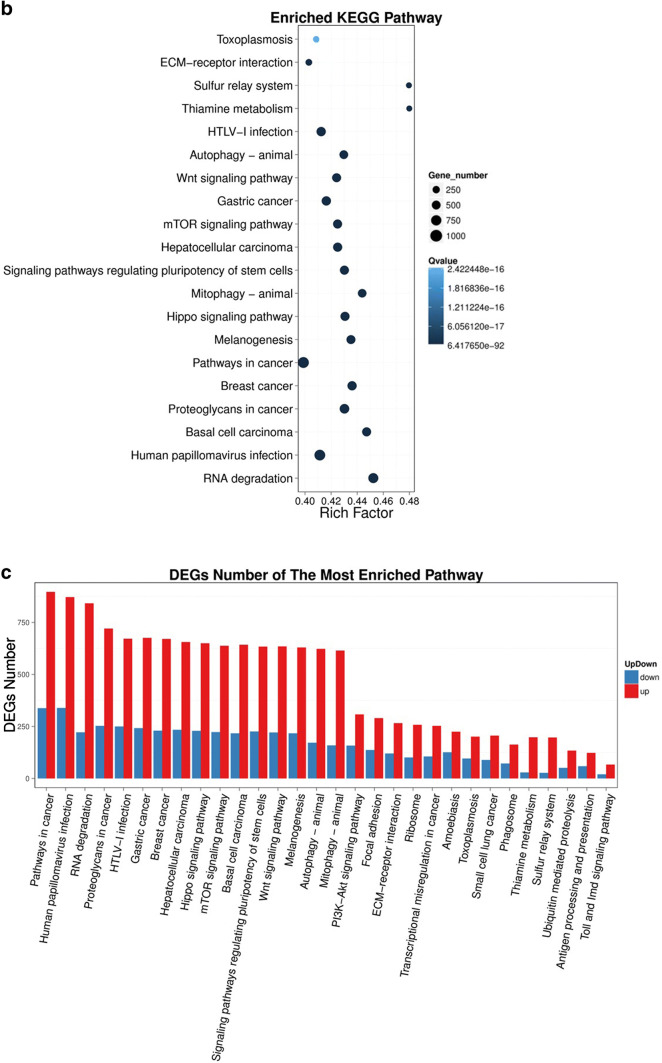
To examine the expression profile of DEGs in our result, DEGs (both upregulated and downregulated) were then subjected to the KEGG pathway enrichment analysis. More than 23% of the DEGs could be annotated. The pathway classification results comparing group NC and group sh1/sh2 are shown in Fig. 3a and supplementary Fig. S2a, and the pathway functional enrichment results are shown in Fig. 3b and supplementary Fig. S2b. The pathway functional enrichment results for up- or down-regulated genes are shown in Fig. 3c and supplementary Fig. S2c. The top ten KEGG pathways with the highest representation of the DEGs are shown in Table 3. We found that most DEGs were enriched in cancer pathways (ko05200), including breast cancer (ko05224), gastric cancer (ko05226) and hepatocellular carcinoma (ko05225), and that some DEGs were also enriched in Wnt (ko04310), mTOR (ko04150) signaling pathways and pathways in regulating pluripotency of stem cells (ko04550).
Fig. 3.
Pathway functional enrichment of DEGs between group NC and group sh1. a Pathway classification of DEGs; b Pathway functional enrichment of DEGs. c Pathway functional enrichment results for up- or down-regulated genes. X axis represents the term of pathways. Y axis represents the number of up- or down-regulated genes
Table 3.
Top ten KEGG pathways with a high representation of DEGs
| Pathway | DEGs genes with pathway annotation (5617) | All genes with pathway annotation (23,480) | p value | q value | Pathway ID |
|---|---|---|---|---|---|
| Pathways in cancer | 1235 (19.8%) | 3097 (13.19%) | 8.962456e-68 | 4.959226e-66 | ko05200 |
| Melanogenesis | 847 (13.58%) | 1946 (8.29%) | 3.684796e-64 | 1.595685e-62 | ko04916 |
| Wnt signaling pathway | 856 (13.72%) | 2018 (8.59%) | 8.728826e-59 | 2.069979e-57 | ko04310 |
| Breast cancer | 901 (14.44%) | 2066 (8.8%) | 4.650451e-69 | 3.087899e-67 | ko05224 |
| Gastric cancer | 918 (14.72%) | 2205 (9.39%) | 8.712956e-59 | 2.069979e-57 | ko05226 |
| Proteoglycans in cancer | 974 (15.61%) | 2264 (9.64%) | 2.275643e-71 | 1.888784e-69 | ko05205 |
| Hepatocellular carcinoma | 890 (14.27%) | 2094 (8.92%) | 9.088525e-62 | 2.743082e-60 | ko05225 |
| Hippo signaling pathway | 879 (14.09%) | 2041 (8.69%) | 3.845024e-64 | 1.595685e-62 | ko04390 |
| Signaling pathways regulating pluripotency of stem cells | 860 (13.79%) | 1999 (8.51%) | 2.096168e-62 | 6.959278e-61 | ko04550 |
| mTOR signaling pathway | 861 (13.8%) | 2026 (8.63%) | 1.376305e-59 | 3.807777e-58 | ko04150 |
Discussion
TOX is aberrantly overexpressed in CTCLs, such as MF and SS. Stable knockdown of TOX in CTCL cells reduces cell cycle progression and promotes apoptosis, leading to inhibited cell viability and colony-forming ability in vitro and suppressed tumor growth in vivo [12]. After TOX gene knockdown, many genes are highly expressed, such as two cyclin-dependent kinase inhibitors (CDKNs), including CDKN1B and CDKN1C) [15]. It has been reported that TOX is able to regulate cell cycle in primary Sézary cells and cutaneous T cell lymphoma, whereas TOX knockdown leads to cell cycle arrest and secondary cell death [12, 16]. In our study, we found that, after TOX knockdown, some proliferation and apoptosis-associated genes, such as PFKFB3, CDK5 and CKKN2A, were up- or down-regulated and most DEGs were enriched in cellular process and cancer pathways, which highlights the importance of TOX in cancer process. As we noted, both changes in apoptosis and cell cycle characteristics of the groups after gene knockdown, it is not clear if the differentially expressed genes are directly the result of interactions with TOX or the result of downstream cell cycle-dependent changes.
HOX genes, including HOXC9, HOXC4, HOXC5, HOXC8, HOXC10, HOXC11 and HOXC13, were significantly downregulated after TOX knockdown, with HOXC9 being downregulated to the highest degree. HOX genes are homeobox genes that function as transcription factors. In humans, 39 HOX genes have been assigned to 13 paralogous groups in four separate clusters termed HOXA, HOXB, HOXC and HOXD [8]. HOXC9 is aberrantly expressed in breast cancer, lung cancer, body fat mass and astrocytoma [3, 8, 14, 22]. HOXC9 can induce neuronal differentiation of neuroblastoma cells [26]. Wang et al. [25] demonstrate that HOXC9 can directly regulate distinct sets of genes to coordinate diverse cellular processes during neuronal differentiation. This may explain why TOX knockdown will lead to less cell viability and colony-forming ability in vitro and reduce tumor growth in vivo.
Through DEGs GO analysis, we found most DEGs are related to the cellular process, biological regulation and binding process. This can explain why TOX knockdown will induce inhibited cell viability as previously reported [7]. With DEGs pathway analysis applied in KEGG, we find two important tumor-related pathways, Wnt and mTOR. They are generally associated with cellular proliferation, differentiation and apoptosis in invertebrates and mammals [4]. β-catenin is expressed by tumor cells in cutaneous lymphoproliferative disorders at various frequencies, and activation and accumulation of β-catenin plays an important role in the development of skin lymphomas [1]. CTCL cells display mTORC1 activation in the lymphoma stage-related fashion with the highest percentage of positive cells identified at the late stage [17]. Treatment with rapamycin can persistently inhibit mTORC1 signaling, and the combined inhibition of mTORC1 and MNK could totally abrogate the growth of CTCL cells [18]. Taking together, these findings could help to understand the mechanism of action of TOX in CTCL and provide clues to novel therapeutics for CTCL.
Several strategies have been employed to enhance the efficacy of current treatments and to find new therapeutic options to improve survival and quality of life for patients with SS and other forms of advanced CTCL [19, 20, 30]. TOX encodes a high-mobility group family (HMG) domain binding nuclear protein which regulates the differentiation of developing T cells. It is thought of as a molecular marker for histological diagnosis of CTCL [6, 31]. Our work has addressed the role of DEGs after TOX knockdown, as GO functional enrichment and pathway analysis have indicated. A limitation of this work is that findings so far are restricted to a single cell line. However, we believe the results may provide some insights into the mechanism of TOX in CTCL as well as candidate targets for therapy of CTCL in the near future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This project was sponsored by the National Natural Science Foundation of China (Grant Number 81602397) and the Natural Science Foundation of Shanghai (Grant Number 15ZR1405700). We thank Prof. Jan Peter Dutz, from the Department of Dermatology and Skin Science, University of British Columbia, Canada, for his critical comments. We are grateful to Prof. Qingfeng Wu, from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for his kind guidance on the statistical analysis of RNA sequencing (RNAseq).
Funding
This project was sponsored by the National Natural Science Foundation of China (Grant Number 81602397) and the Natural Science Foundation of Shanghai (Grant Number 15ZR1405700).
Compliance with ethical standards
Conflict of interest
The authors state no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bellei B, Pacchiarotti A, Perez M, Faraggiana T. Frequent beta-catenin overexpression without exon 3 mutation in cutaneous lymphomas. Mod Pathol. 2004;17:1275–1281. doi: 10.1038/modpathol.3800181. [DOI] [PubMed] [Google Scholar]
- 2.Boonk SE, Cetinozman F, Vermeer MH, Jansen PM, Willemze R. Differential expression of TOX by skin-infiltrating T cells in Sezary syndrome and erythrodermic dermatitis. J Cutan Pathol. 2015;42:604–609. doi: 10.1111/cup.12490. [DOI] [PubMed] [Google Scholar]
- 3.Brune JE, Kern M, Kunath A, et al. Fat depot-specific expression of HOXC9 and HOXC10 may contribute to adverse fat distribution and related metabolic traits. Obesity (Silver Spring) 2016;24:51–59. doi: 10.1002/oby.21317. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 5.Dulmage BO, Geskin LJ. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br J Dermatol. 2013;169:1188–1197. doi: 10.1111/bjd.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Litvinov IV, Wang Y, et al. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget. 2014;5:4418–4425. doi: 10.18632/oncotarget.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Su MW, Jiang X, Zhou Y. Evidence of an oncogenic role of aberrant TOX activation in cutaneous T-cell lymphoma. Blood. 2015;125:1435–1443. doi: 10.1182/blood-2014-05-571778. [DOI] [PubMed] [Google Scholar]
- 8.Hur H, Lee JY, Yang S, Kim JM, Park AE, Kim MH. HOXC9 Induces phenotypic switching between proliferation and invasion in breast cancer cells. J Cancer. 2016;7:768–773. doi: 10.7150/jca.13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kari L, Loboda A, Nebozhyn M, et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med. 2003;197:1477–1488. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kioussis D. Thymocyte differentiation: it's time to bend a little. Nat Immunol. 2002;3:214–215. doi: 10.1038/ni0302-214. [DOI] [PubMed] [Google Scholar]
- 11.Klemke CD, Goerdt S, Schrama D, Becker JC. New insights into the molecular biology and targeted therapy of cutaneous T-cell lymphomas. J Dtsch Dermatol Ges. 2006;4:395–406. doi: 10.1111/j.1610-0387.2006.05982.x. [DOI] [PubMed] [Google Scholar]
- 12.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Q, Geng J, Ma K, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135:1675–1684. doi: 10.1007/s00432-009-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litvinov IV, Netchiporouk E, Cordeiro B, et al. The use of transcriptional profiling to improve personalized diagnosis and management of cutaneous T-cell lymphoma (CTCL) Clin Cancer Res. 2015;21:2820–2829. doi: 10.1158/1078-0432.CCR-14-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobbardi R, Pinder J, Martinez-Pastor B, et al. TOX regulates growth, DNA repair, and genomic instability in T-cell acute lymphoblastic leukemia. Cancer Discov. 2017;7:1336–1353. doi: 10.1158/2159-8290.CD-17-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzec M, Liu X, Kasprzycka M, et al. IL-2- and IL-15-induced activation of the rapamycin-sensitive mTORC1 pathway in malignant CD4+ T lymphocytes. Blood. 2008;111:2181–2189. doi: 10.1182/blood-2007-06-095182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzec M, Liu X, Wysocka M, Rook AH, Odum N, Wasik MA. Simultaneous inhibition of mTOR-containing complex 1 (mTORC1) and MNK induces apoptosis of cutaneous T-cell lymphoma (CTCL) cells. PLoS ONE. 2011;6:e24849. doi: 10.1371/journal.pone.0024849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.McGirt LY, Adams CM, Baerenwald DA, Zwerner JP, Zic JA, Eischen CM. miR-223 regulates cell growth and targets proto-oncogenes in mycosis fungoides/cutaneous T-cell lymphoma. J Invest Dermatol. 2014;134:1101–1107. doi: 10.1038/jid.2013.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morimura S, Sugaya M, Suga H, et al. TOX expression in different subtypes of cutaneous lymphoma. Arch Dermatol Res. 2014;306:843–849. doi: 10.1007/s00403-014-1501-7. [DOI] [PubMed] [Google Scholar]
- 21.Nihal M, Ahmad N, Wood GS. SIRT1 is upregulated in cutaneous T-cell lymphoma, and its inhibition induces growth arrest and apoptosis. Cell Cycle. 2014;13:632–640. doi: 10.4161/cc.27523. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto OK, Oba-Shinjo SM, Lopes L, Nagahashi Marie SK. Expression of HOXC9 and E2F2 are up-regulated in CD133(+) cells isolated from human astrocytomas and associate with transformation of human astrocytes. Biochim Biophys Acta. 2007;1769:437–442. doi: 10.1016/j.bbaexp.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Olsen E, Vonderheid E, Pimpinelli N, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC) Blood. 2007;110:1713–1722. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 24.Schrader AMR, Jansen PM, Willemze R. TOX expression in cutaneous T-cell lymphomas: an adjunctive diagnostic marker that is not tumour specific and not restricted to the CD4(+) CD8(−) phenotype. Br J of Dermatol. 2016;175:382–386. doi: 10.1111/bjd.14508. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Choi JH, Ding J, et al. HOXC9 directly regulates distinct sets of genes to coordinate diverse cellular processes during neuronal differentiation. BMC Genomics. 2013;14:830. doi: 10.1186/1471-2164-14-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Yang L, Choi JH, et al. Genome-wide analysis of HOXC9-induced neuronal differentiation of neuroblastoma cells. Genom Data. 2014;2:50–52. doi: 10.1016/j.gdata.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson B, Chen JYF, Han P, Rufner KM, Goularte OD, Kaye J. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 28.Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105:3768–3785. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Luo Y, Liu J, Liu Y, Sun Q. TOX acts an oncological role in mycosis fungoides. PLoS ONE. 2015;10:e0117479. doi: 10.1371/journal.pone.0117479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Zhang X, Zhu H, Wu X, et al. A genetic polymorphism in TOX3 is associated with survival of gastric cancer in a Chinese population. PLoS ONE. 2013;8:e72186. doi: 10.1371/journal.pone.0072186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Wang Y, Yu R, et al. Molecular markers of early-stage mycosis fungoides. J Invest Dermatol. 2012;132:1698–1706. doi: 10.1038/jid.2012.13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.