Fig.6. Genes in the 16-gene ACS signature that mediate detrimental effects during infection.
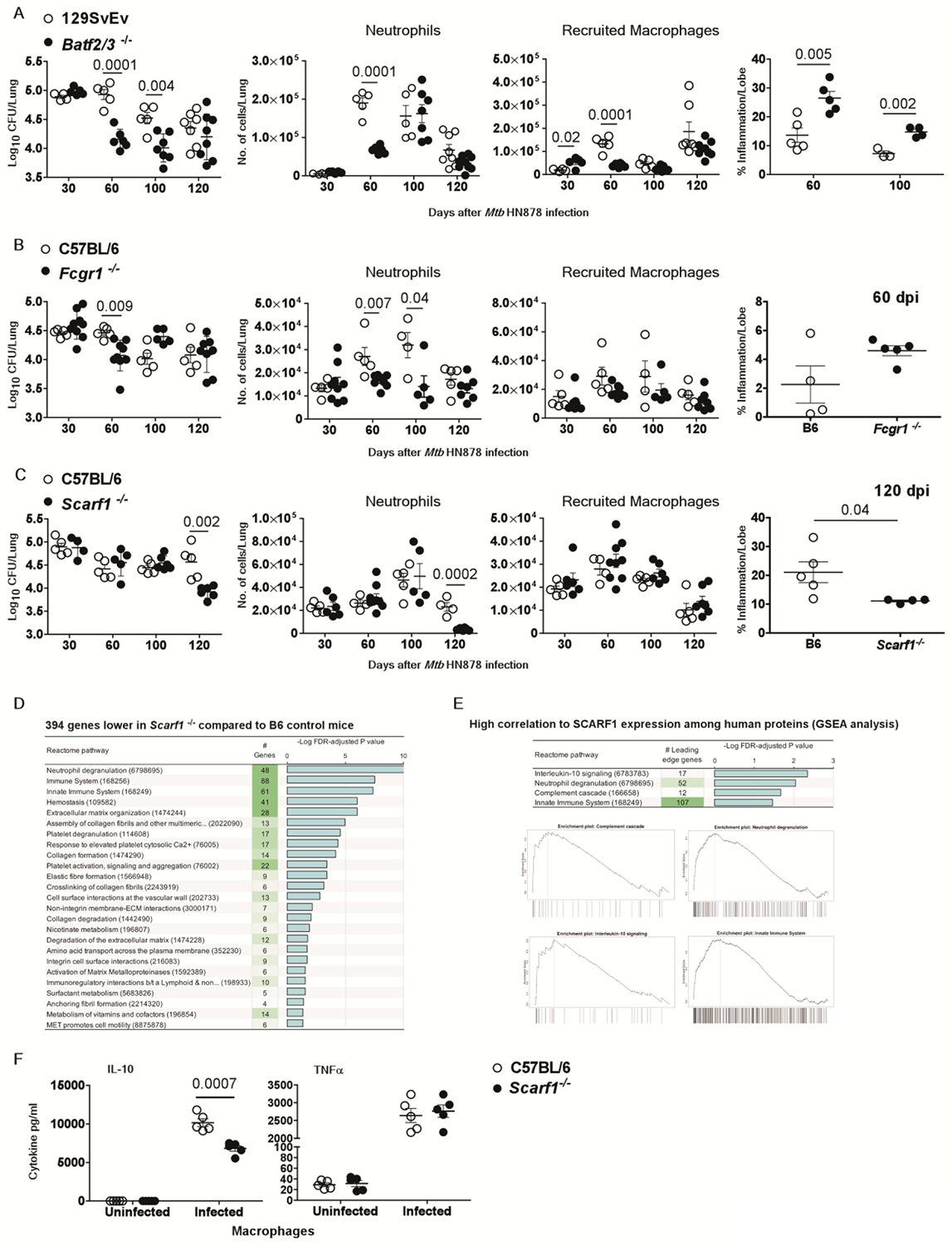
Wild-type or gene-deficient mice strains were infected with Mtb HN878 (100 CFU) by aerosol route. On 30, 60, 100 and 120 days post-infection (dpi), lungs were harvested to assess bacterial burden by plating, or inflammatory cell infiltration by flow cytometry, or histologically by H&E staining in (A) Batf2/3r−/− mice, (B) Fcgrt−/− mice (C) Scarft−/− mice and relevant controls (D) DESeq2 differential gene expression comparisons identified in infected Scarft−/− mice compared to wild-type mice at 120 dpi respectively, (E) Gene set enrichment analysis (GSEA) for Reactome pathways significantly enriched among human proteins with high correlation to SCARF1. Enrichment plots are shown for each of the enriched terms, showing the relative rank of genes from the enriched pathways (across the x-axis) and (F) IL-10 and TNFa production was measured in culture supernatants from uninfected and Mtb HN878-infected bone marrow derived macrophages (MOI 1) from C57BL/6J and ScarfT−/− mice after 2 dpi. The data points represent the means (±SEM) of one out of two separate experiments. Bacterial burden in gene deficient mice compared to control mice and cytokine production from macrophages was done by unpaired two-tailed Student’s t-test at different dpi.
