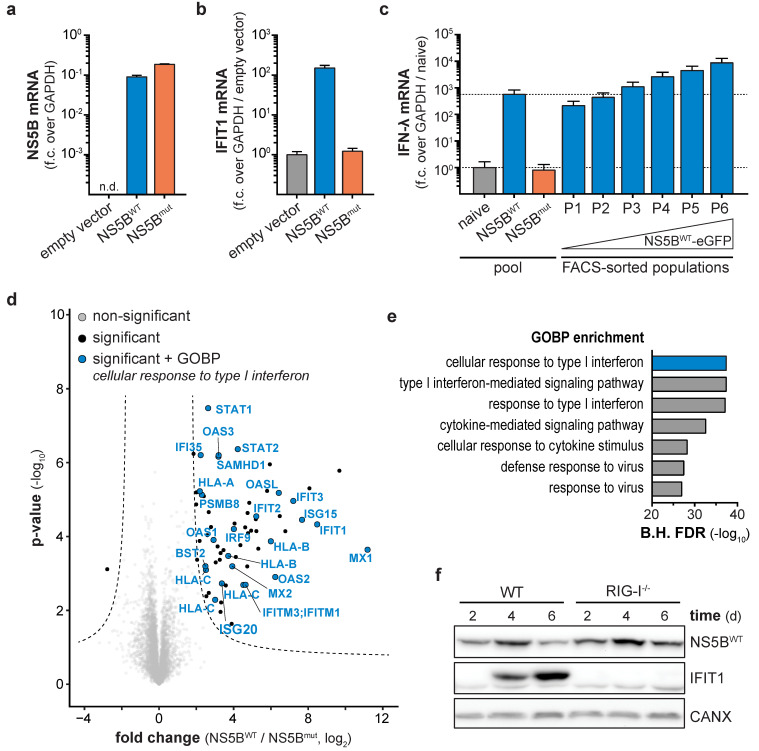
Figure 1.
Ligand-based stimulation of retinoic acid inducible gene I (RIG-I) by expression of Hepatitis C virus (HCV) RdRP NS5B. NS5B (a) and IFIT1 (b) mRNA levels in NS5BWT, NS5Bmut, or empty vector transduced A549 cells, 6 days post transduction. NS5B fold change (f.c.) over GAPDH or IFIT1 fold change over GAPDH and empty vector control was calculated (mean + s.d. of n = 3 technical replicates). One representative of n = 3 independent experiments is shown. (c) IFN-λ mRNA levels in naïve cells or cells transduced with NS5BWT or NS5Bmut either correspond to the entire cell pool or FACS sub-populations (P1-6), which were sorted by increasing NS5BWT-eGFP levels. Fold change over GAPDH and naïve cells was calculated (mean + s.d. of n = 3 technical replicates). Dashed lines indicate IFIT1 mRNA levels of the cell pool for naïve or NS5BWT transduced cells. (d) Total proteome analyses of A549 cells 12 days post NS5BWT or NS5Bmut transduction. Hyperbolic lines indicate significantly changing proteins (two-tailed Student’s t-test, S0 = 1, permutation-based FDR < 0.01, n = 4 technical replicates). Proteins annotated with the GOBP term “cellular response to type I interferon” are highlighted in blue. (e) Bar plot showing Benjamini–Hochberg-adjusted (B.H.) FDR values (Fisher’s exact test) for the seven most significantly enriched GOBP terms within significantly changing proteins upon NS5BWT expression. (f) Western blot of NS5B and IFIT1 protein expression in wild-type and RIG-I knock-out A549 cells transduced with NS5BWT for the indicated days. Calnexin (CANX) was used as loading control.