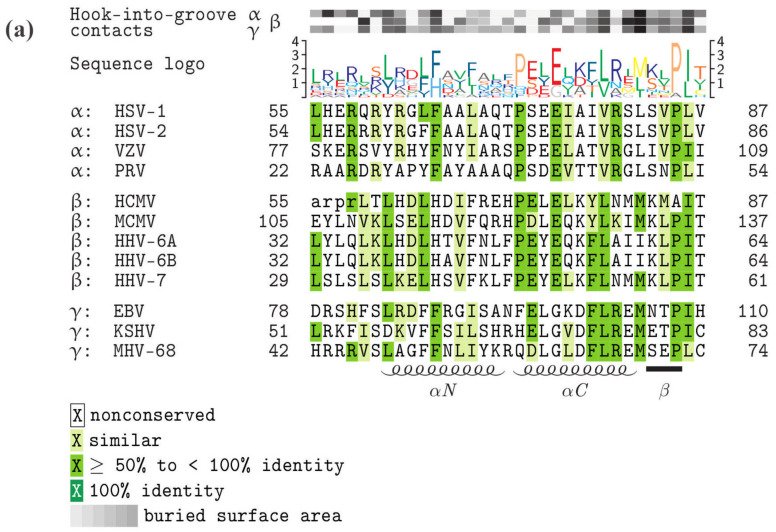
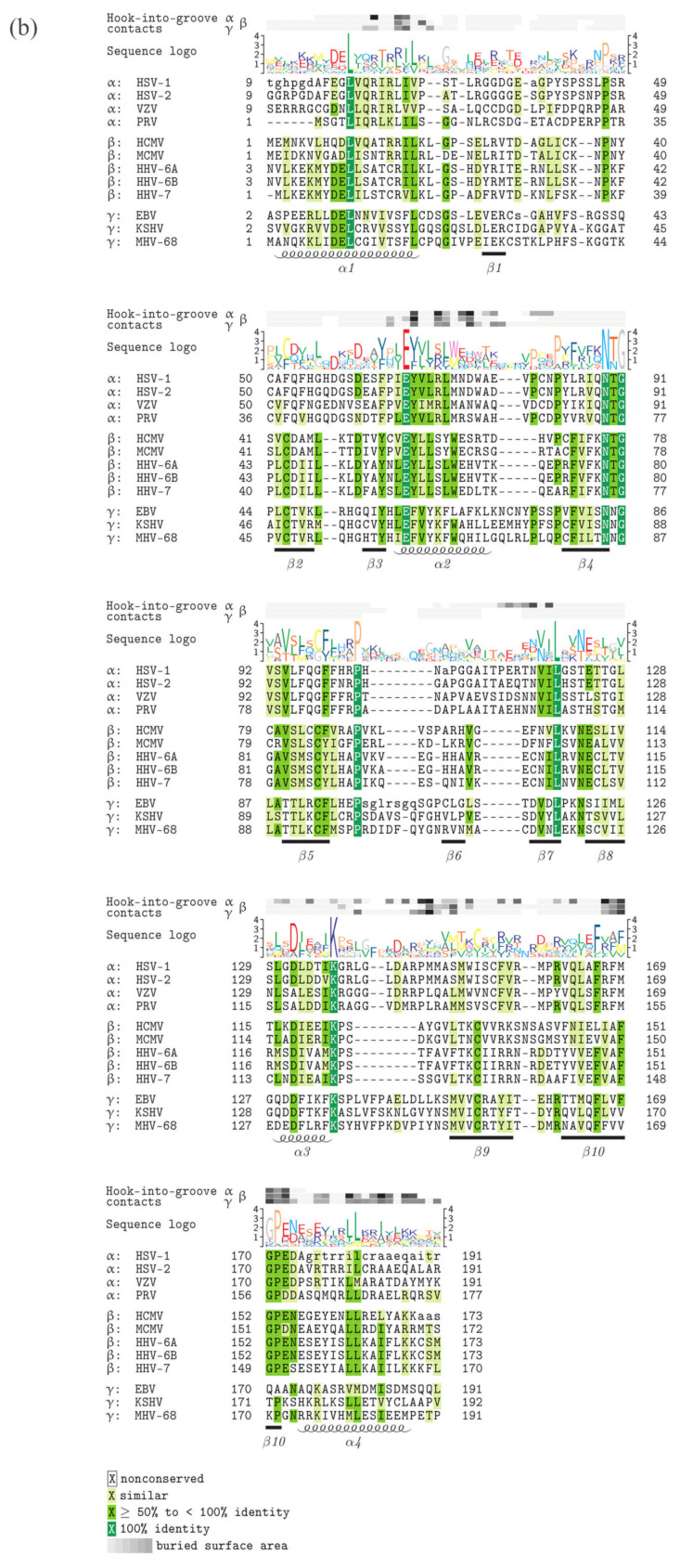
Figure 2.
Core NEC protein sequence alignments, highlighting the functionally important hook and groove domains, conserved amino acids and amino acids representing contact interfaces. (a) Homologs of HCMV pUL53 (hook proteins), (b) homologs of HCMV pUL50 (groove proteins). Increasing levels of sequence conservation are indicated by darker shades of green in the alignment and by higher letters in the sequence logo above the alignment. Lower-case letters mark those residues that were not resolved in the crystal structures of the respective proteins. The buried surface area at hook-into-groove contact positions in α-, β-, and γ-herpesviruses is indicated by gray squares. Darker shades of gray indicate a larger buried surface. The elements of secondary structure are depicted schematically below the alignment. GenbankTM accession numbers: HSV-1, P10218 and P10215; HSV-2, P89457 and P89454; VZV, P09280 and P09283; PRV (SuHV-1), T2FKZ7 and G3G955; HCMV, P16791 and P16794; MCMV (MuHV-1), D3XDN8 and D3XDP1; HHV-6A, P52465 and P28865; HHV-6B, Q9QJ35 and Q9WT27; HHV-7, P52466 and P52361; EBV, P03185 and P0CK47; KSHV (HHV-8), F5HA27 and F5H982; MHV-68 (MuHV-4), O41968 and O41970.