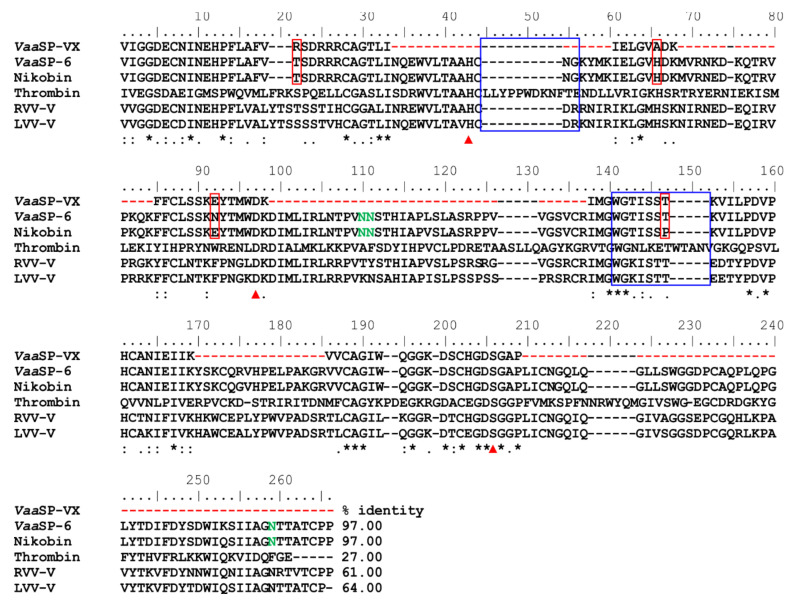
Figure 5.
Partial amino acid sequence of VaaSP-VX and its comparison with sequences of some relevant serine proteases (SPs). VaaSP-VX was sequenced using a combination of Edman degradation and tandem MS. Based on its established partial structure, VaaSP-VX was recognized as an isoform of VaaSP-6, whose complete cDNA sequence was determined and deposited at the National Center for Biotechnology Information (NCBI, MG958495). Nikobin, an SP isolated from Vipera nikolskii venom, is a protein with the same extent of structural identity to the sequenced parts of VaaSP-VX as VaaSP-6 (97.0%). Amino acid residues, where differences between VaaSP-VX and VaaSP-6 or nikobin occurred, are in red boxes. Red arrows at the bottom of the aligned sequences point to three amino acid residues that constituted the catalytic triad in SPs (His57, Asp102, and Ser195, according to chymotrypsin numbering). The undetermined parts of the VaaSP-VX sequence are denoted by red dashes, while predicted gaps are denoted by black ones. Amino acid residues that may be N-glycosylated in VaaSP-VX (if present) are printed in green. Other SPs that were aligned are human thrombin, which is the main physiological activator of FV, and two snake venom FV activators, namely RVV-V from Daboia siamensis and LVV-V from Macrovipera lebetina. Except for thrombin, the two loops, 44-loop and 148-loop (in blue squares), are absent in all presented FV activators from snake venoms. This was established as the reason why snake venom SPs were resistant to inhibition by antithrombin. Below the sequences, an asterisk (*) designates a position that is strictly conserved in all aligned sequences, a colon (:) denotes a position in which only conservative substitutions were detected, and a dot (.) denotes a position harboring semi-conservative substitutions. On all other sites, the substitutions were non-conservative. The percentages of identity between the sequence of VaaSP-VX and the corresponding sequences in VaaSP-6, nikobin, thrombin, RVV-V, and LVV-V are presented.