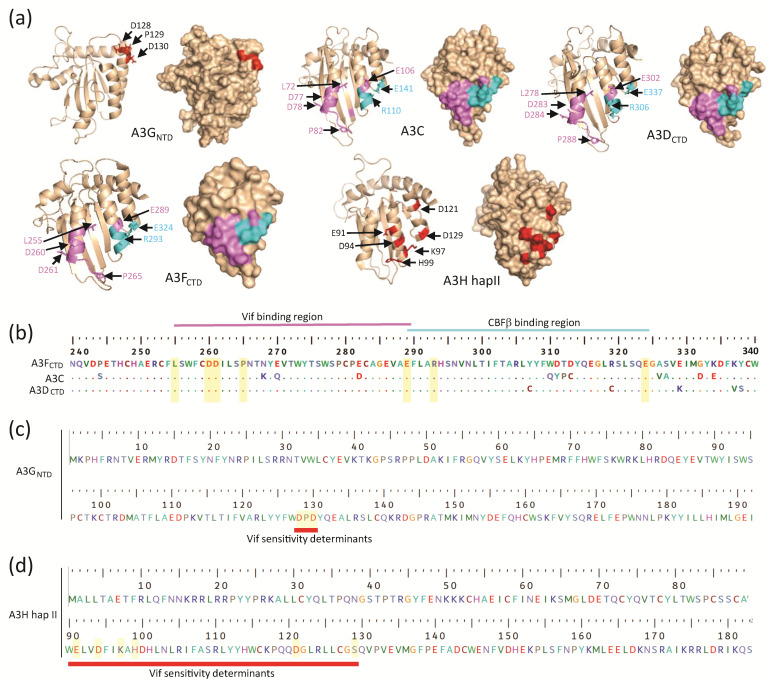
Figure 3.
Comparison of A3GNTD, A3C, A3DCTD, A3FCTD, and A3H hap II structures. (a) Cartoon and surface representations of the structure of A3GNTD (PDB: 2MZZ), A3C (PDB: 3VOW), and A3DCTD (a predicted model generated through homology modeling based on the A3FCTD structure (PDB: 6NIL) by using the Phyre2 web portal for protein modeling, prediction and analysis [140], and A3H hap II (PDB: 6BBO). The known Vif-mediated degradation determinants labeled in red (for A3G and A3H), the critical residues for Vif-binding in magenta (for A3C, A3D, and A3F), and the CBFβ binding residues in cyan (for A3C, A3D, A3F). (b) Multiple sequence alignment of A3 domains (A3FCTD, A3C, A3DCTD) and the critical Vif- and CBFβ-binding residues are highlighted in yellow bars. A3F was used as a reference and hence the numbering is based on A3F’s residue numbers. The determinants for Vif- and CBFβ-binding are in non-overlapping regions, which are indicated above the alignment. (c,d) Similar primary sequences of residues in A3GNTD and A3H hap II are shown, respectively. The residues highlighted in yellow bars are identified to be critical for Vif-mediated degradation of the respective A3 proteins. The sequence alignment was performed by a Clustal W multiple alignment in BioEdit.