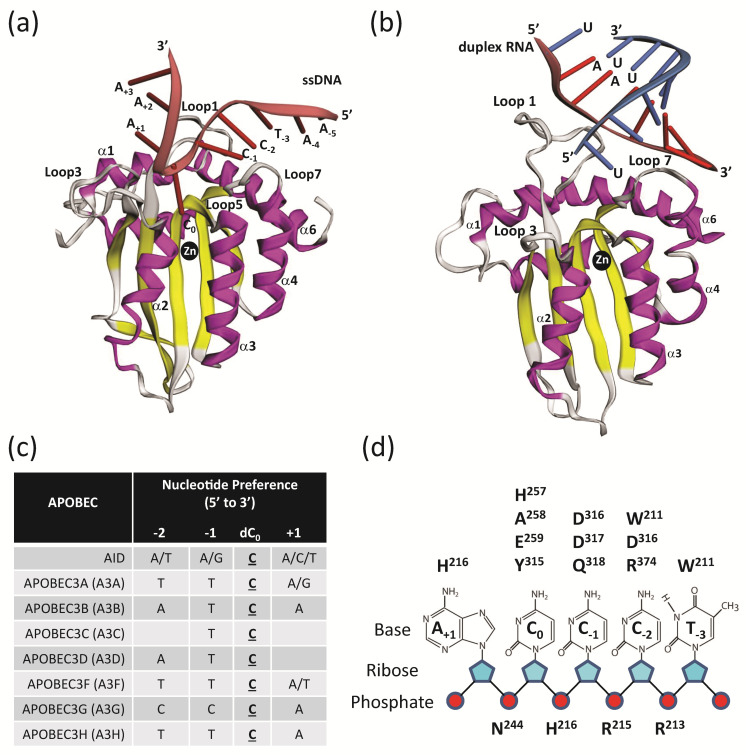
Figure 6.
A3 deamination and ssDNA interactions. (a) Co-crystal structure of A3GCTD in complex with ssDNA substrate 5′-AATCCCAAA-3′ (PDB 6BUX). Shown is the three-dimensional fold of the A3GCTD with α-helices (purple) and β-sheets (yellow) with the ssDNA (red) co-ordinated with the reactive cytosine (C0) positioned next to the catalytic zinc atom. Loops 1, 3, 5, and 7, which influence substrate binding, specificity, and dinucleotide preference selection, are labeled. (b) Co-crystal of A3H in complex with duplex RNA substrate 5′-UAAAAAAA-3′ + 5′-UUUUUUUUU-3′ with a 1 nucleotide overhang (PDB 6B0B). Shown are α-helices (purple) and β-sheets (yellow) with the duplex RNA (red and blue) positioned with zinc atom. (c) Preferred ssDNA substrates of the A3 enzymes where the underlined C is the preferred target nucleotide of the deamination reaction. (d) Summary of the interactions of the 5′-TCCCA target ssDNA substrate with amino acids in A3GCTD (PBD 6BUX). A3G loop 1 contains amino acids W211, R213, R215, H216; A3G loop 3 contains amino acids N244, H257; A3G α-helix 2 contains amino acids A258 and E259; A3G loop 7 contains amino acids Y315, D316, D317, Q318 and A3G α-helix 6 contains amino acid R374.