Figure 4.
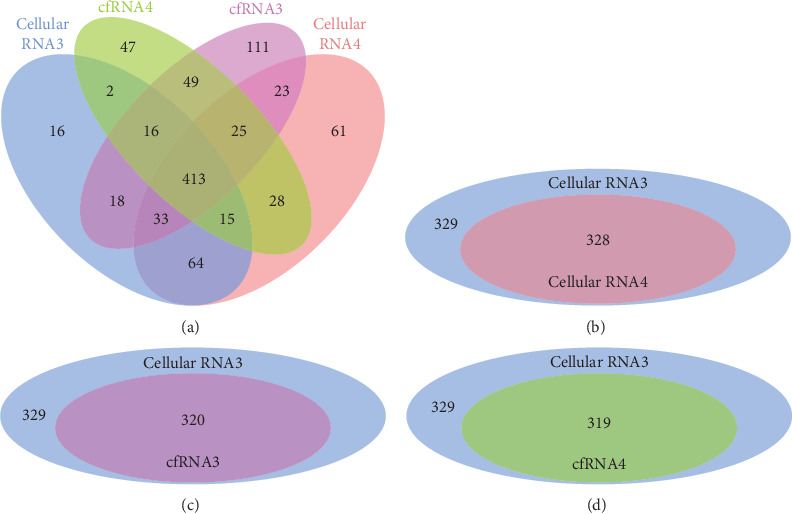
The BM supernatant contains stable miRNAs with as good integrity as in BM cells. (a) Cellular RNA (from cells in fresh BM of donors 3 and 4) and cfRNA (from fresh BM of donor 4 and RT-7-days BM of donor 3) were used to prepare the miRNA NGS library and sequence. After the bioinformatics analysis, detectable known human miRNAs based on miRBase 22.0 in different samples were compared. The graph shows the number of identical and different miRNAs between samples. To avoid the experimental bias in lower sequencing read data, detected miRNAs in cellular RNA3 whose normalized reads were greater than 50 were identified as a base pool; then, miRNAs in the base pool were compared with detectable miRNAs in other samples. The graphs show the numbers of identical miRNAs in the base pool and detectable miRNA in cellular (b) RNA4, (c) cfRNA3, and (d) cfRNA4.
