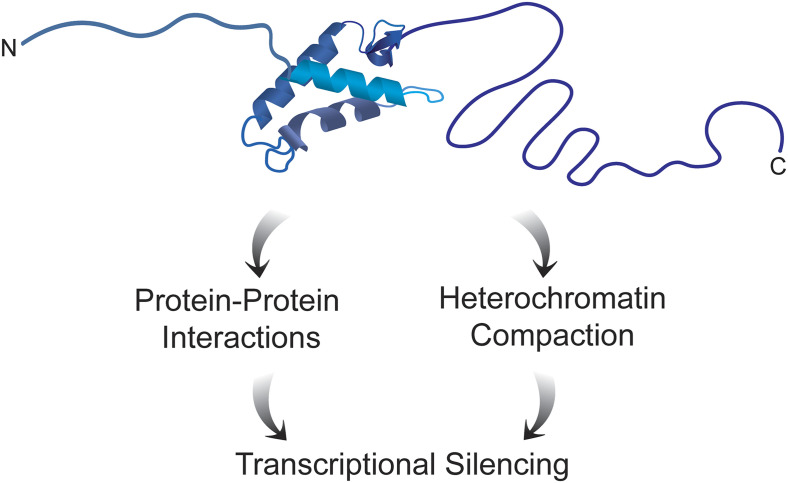
Eukaryotic genomes at their core consist of a nucleoprotein complex termed chromatin. The subunit of chromatin is the nucleosome, which is formed from 146 bp of DNA wrapped around an octamer of core histone proteins (H2A, H2B, H3, and H4). An array of nucleosomes connected by intervening linker DNA segments represents the chromatin fiber in its simplest form. In reality, the chromatin fiber that encompasses a given region of the genome is associated with a distinctive set of proteins that specify the functionality of that chromatin region. For example, the euchromatin of an active gene will be bound to transcription factors, chromatin remodelers, and chromatin-modifying enzymes. On the other hand, a gene that is transcriptionally silenced by constitutive heterochromatin will be assembled with nucleosomes marked by histone H3 lysine 9 trimethylation (H3K9me3) and bound to HP1 and other proteins. While the connection between constitutive heterochromatin and transcriptional silencing is well established, how silencing is achieved is not well understood. In PNAS, Healton et al. (1) show that linker histone H1 is enriched in the constitutive heterochromatin that silences repetitive elements in mouse embryonic stem cells (mESCs), and that acute depletion of H1 leads to substantial derepression of repetitive element gene expression. Surprisingly, H1 exerts its effects through two fundamentally different mechanisms, one involving H1–protein interactions and the other involving modulation of the higher-order structure of heterochromatin domains (Fig. 1). These results raise new questions regarding how linker histones function in chromatin.
Fig. 1.
Schematic structure of linker histone H1 drawn to approximate scale. The work of Healton et al. (1) describes two different mechanisms used by H1 to silence heterochromatin, protein–protein interactions, and global chromatin compaction.
Linker histones are the most abundant chromatin-associated proteins in most eukaryotic genomes, with an average of 0.5 to 1.3 H1 per nucleosome depending on cell type (2). There are seven H1 sequence variants in somatic cells. All variants share the same general structure shown in Fig. 1. The central ∼80 residues fold into a globular winged helix motif while the ∼35-residue N-terminal domain and the long ∼100-residue C-terminal domain (CTD) are disordered. H1 binds to the nucleosome via its winged helix domain. Previous studies from the Skoultchi laboratory have shown that linker histones are required for proper development in mice (3). If the genes for one or two variants are knocked out, normal nuclear H1 levels are maintained due to increased expression of the remaining variants (4). However, when three variant genes (H1c/d/e) are knocked out, mice display embryonic lethality (3).
Although ubiquitous, the distribution of linker histones throughout the genome is not uniform. For instance, linker histone levels are reduced in the chromatin that encompasses transcriptionally active gene promoters (5, 6). In contrast, Healton et al. (1) were interested in identifying those regions of the genome that were enriched in linker histones. They used the ISOR algorithm to analyze available chromatin immunoprecipitation-sequence data for the H1d variant and found that it was significantly enriched in chromatin bearing the H3K9me3 modification, a hallmark of constitutive heterochromatin. Similarly, enrichment of H1 was observed in chromatin bound to the Suv39h1, Suv39h2, and SETDB1 histone methyltransferases that are responsible for trimethylating H3K9. Consistent with previous results (5, 6), H1 was strongly depleted in chromatin bound to the G9a methyltransferase and marked by acetylated H3K9 and methylated H3K4, all signatures of active euchromatin. The ISOR analyses subsequently were confirmed by several other approaches. Taken together, the computational analyses indicated that H1 was enriched in constitutive heterochromatin domains throughout the genome.
Repetitive DNA elements make up about 50% of the genomes of mice and humans. These sequences include satellite DNA, long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and endogenous retroviruses (ERVs). In mESCs, repetitive elements are silenced by constitutive heterochromatin marked by H3K9me3. Perhaps not surprisingly, the ISOR analysis by Healton et al. (1) also revealed that LINEs, SINEs, ERVs, and pericentromeric satellite sequences were enriched in genomic domains that also were enriched in H1, a result confirmed experimentally. This led Healton et al. (1) to next ask whether H1 had a repressive effect on repetitive element gene expression. The transcript levels of specific members of the major satellite, LINE, and ERV families were examined, initially in H1 triple-knockout mESCs. When compared against control cells, small increases in transcript levels were sometimes observed upon H1 depletion. However, the total H1 content in triple-knockout mESCs is only decreased by 50% compared to wild type because of up-regulation of the remaining H1a and H1b genes. To get around this problem, Healton et al. (1) used CRISPR-Cas9 technology to delete both copies of the H1b gene and one copy of the H1a gene present in triple-knockout mESCs. This reduced the total H1 content to only 20% of that in wild-type cells. Importantly, in these “H1-low” ESCs the level of major satellite transcripts was increased by 100-fold, and similar derepressive effects were seen with LINE-1 and ERV transcripts. These results demonstrate that H1 is a major contributor to transcriptional silencing by constitutive heterochromatin in mESCs.
The involvement of H1 in H3K9me3-mediated heterochromatin silencing suggests a possible relationship between H1 and the enzymes that lay down the H3K9me3 mark. In support of this idea, previous studies from the Skoultchi laboratory showed that Drosophila H1 binds to Su(var)3-9, the fly ortholog of mammalian Suv39h1/h2 (7). The H1–enzyme interaction promotes H3K9me3 deposition and transcriptional repression of repetitive DNA in flies. Healton et al. (1) performed a number of experiments to determine if similar structure/function relationships exist in the mouse. GST pulldowns were used to demonstrate physical interaction between six of the somatic isoforms and Suv39h1/2. Further, truncation of the H1d CTD disrupted its interaction with Suv39h1/2 and SETDB1, indicating that CTD mediates these protein–protein interactions. The next question addressed was whether H1 stimulated methyltransferase enzymatic activity in a chromatin context in vitro. Recombinant dinucleosomes reconstituted with and without H1 were used as substrates for Suv39h1, SETDB1, and G9a. Results indicated that H3K9 of dinucleosomes bound to H1 was methylated by Suv39h1 and SETDB1 to a much greater extent than the H3K9 of control dinucleosomes. H1-dependent stimulation was specific for the heterochromatin-associated methyltransferases. The in vitro studies indicate that H1 uses its CTD to bind to Suv39h1/2 and SETDB1. Moreover, the CTD–enzyme interaction appears to occur when H1 is simultaneously bound to the nucleosome, leading to local H3K9 methylation. Altogether, the story that has emerged thus far is that H1 silences repetitive elements in part by binding to and stimulating the activity of Suv39h1/2 and SETDB1, thereby promoting H3K9me3 deposition and subsequent assembly of the repetitive DNA into constitutive heterochromatin. These results highlight an underappreciated molecular mechanism through which H1 acts: protein–protein interactions. Although the dogma is that H1 is a DNA- and nucleosome-binding protein, reports of H1–protein interactions have accumulated in the literature over the last 20 y (8). The Skoultchi laboratory now has shown that theDNA methyltransferases
In PNAS, Healton et al. show that linker histone H1 is enriched in the constitutive heterochromatin that silences repetitive elements in mouse embryonic stem cells (mESCs), and that acute depletion of H1 leads to substantial derepression of repetitive element gene expression.
DNMT1 and DNMT3B (9), Drosophila Su(var)3-9 (7), and mouse Suv39h1/h2 and SETDB1 (1) all interact with H1 through the disordered CTD. These studies stand out because they link H1–protein interactions to functional outcomes. In view of these results, studying the prevalence and functional importance of H1–protein interactions should be an interesting and productive area of future research in genome biology.
H1–histone methyltransferase interactions, while important, were not the only mechanism involved in H1-dependent transcriptional silencing of repetitive elements. Healton et al. (1) observed that the level of major satellite expression was much greater in H1-low mESCs compared to cells lacking Suv39h1/2, which suggests that H1-dependent mechanisms other than H3K9me3 deposition exist for silencing repetitive elements. The involvement of H1 in two other potential repressive mechanisms, DNA methylation and H3K27 methylation, were ruled out by present or past experiments. The final parameter examined was chromatin structure. Mild micrococcal nuclease (MNase) digestion is an assay for accessibility of the linker DNA within condensed chromatin. When nuclei from wild-type, Suv39h1/2 double-knockout, and H1-low mESCs were lightly digested with MNase, the amount of satellite DNA released was significantly greater in the H1-low cells than in wild-type cells and those lacking H3K9me3. This suggests that the heterochromatin encompassing the satellite repeats was less condensed when H1 was severely depleted. The same conclusion was obtained using satellite-specific transcription activator-like effector nucleases and by determining the effect of curaxin on major satellite expression. While all three of these experimental approaches suggest a less compact state of pericentric heterochromatin in the absence of H1, they say nothing about what that state is at the structural level.
It is well established that the chromatin fiber equilibrates between unfolded 10-nm “beads-on-a-string” and extensively folded helical 30-nm states in vitro, and that H1 stabilizes the 30-nm state (10). The folded 30-nm state is repressive to transcription (11). Seemingly this is the end of the story—H1 represses gene expression through formation of stable 30-nm chromatin fibers. The problem with this model is that the widespread existence of 30-nm fibers in the nucleus has never been observed. On the contrary, a great deal of evidence suggests that the bulk of the genome is built from long-range interaction of unfolded 10-nm fibers (12, 13). Thus, mechanisms other than formation of 30-nm fibers appear to be involved in linker histone-mediated transcriptional silencing. Interestingly, the chromosomal fiber is in the extended 10-nm state even within heterochromatin domains that are globally condensed (chromocenters) (14). Consistent with this observation, heterochromatin protein 1 (HP1) binds to H3K9me3 and bridges adjacent nucleosomes, leading to a very extended chromatin structure (15). This raises an important question. How is it that the chromatin found within constitutive heterochromatin simultaneously can be locally decondensed and globally compacted? This is where H1 may come in. Recent studies of the condensates formed in vitro by nucleosomal arrays under physiological ionic conditions have shown that H1 stabilizes the interdigitated packaging of the 10-nm arrays within the condensates and makes the condensates smaller and more compact (16). Thus, H1 may condense constitutive heterochromatin by stabilizing long-range interdigitated interaction of locally extended heterochromatin fibers, producing a globally compact state that is repressive to transcription. One word of caution: Mammalian constitutive heterochromatin is a very complex beast consisting of the chromatin fiber, HP1 bound to H3K9me3 and itself, many specific regulatory proteins, and RNA (17), and deciphering the structural basis of its function is not going to be simple. The work of Healton et al. (1) is important in that it raises many questions regarding the role of histone H1 in this puzzle.
Acknowledgments
Research in J.C.H.’s laboratory is supported by National Science Foundation Grant MCB-1814012.
Footnotes
The author declares no competing interest.
See companion article, “H1 linker histones silence repetitive elements by promoting both histone H3K9 methylation and chromatin compaction,” 10.1073/pnas.1920725117.
References
- 1.Healton S. E., et al., H1 linker histones silence repetitive elements by promoting both histone H3K9 methylation and chromatin compaction. Proc. Natl. Acad. Sci. U.S.A. 117, 14251–14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodcock C. L., Skoultchi A. I., Fan Y., Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Fan Y., et al., H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23, 4559–4572 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan Y., Sirotkin A., Russell R. G., Ayala J., Skoultchi A. I., Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol. Cell. Biol. 21, 7933–7943 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izzo A., et al., The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 3, 2142–2154 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Cao K., et al., High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 9, e1003417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X., et al., Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science 340, 78–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalashnikova A. A., Rogge R. A., Hansen J. C., Linker histone H1 and protein-protein interactions. Biochim. Biophys. Acta 1859, 455–461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S.-M., Kim B. J., Norwood Toro L., Skoultchi A. I., H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. U.S.A. 110, 1708–1713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen J. C., Conformational dynamics of the chromatin fiber in solution: Determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31, 361–392 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Tse C., Sera T., Wolffe A. W., Hansen J. C., Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18, 4629–4638 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeshima K., Imai R., Tamura S., Nozaki T., Chromatin as dynamic 10-nm fibers. Chromosoma 123, 225–237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen J. C., et al., The 10-nm chromatin fiber and its relationship to interphase chromosome organization. Biochem. Soc. Trans. 46, 67–76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fussner E., et al., Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 13, 992–996 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machida S., et al., Structural basis of heterochromatin formation by human HP1. Mol. Cell 69, 385–397.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Maeshima K., et al., Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 35, 1115–1132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saksouk N., Simboeck E., Déjardin J., Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]