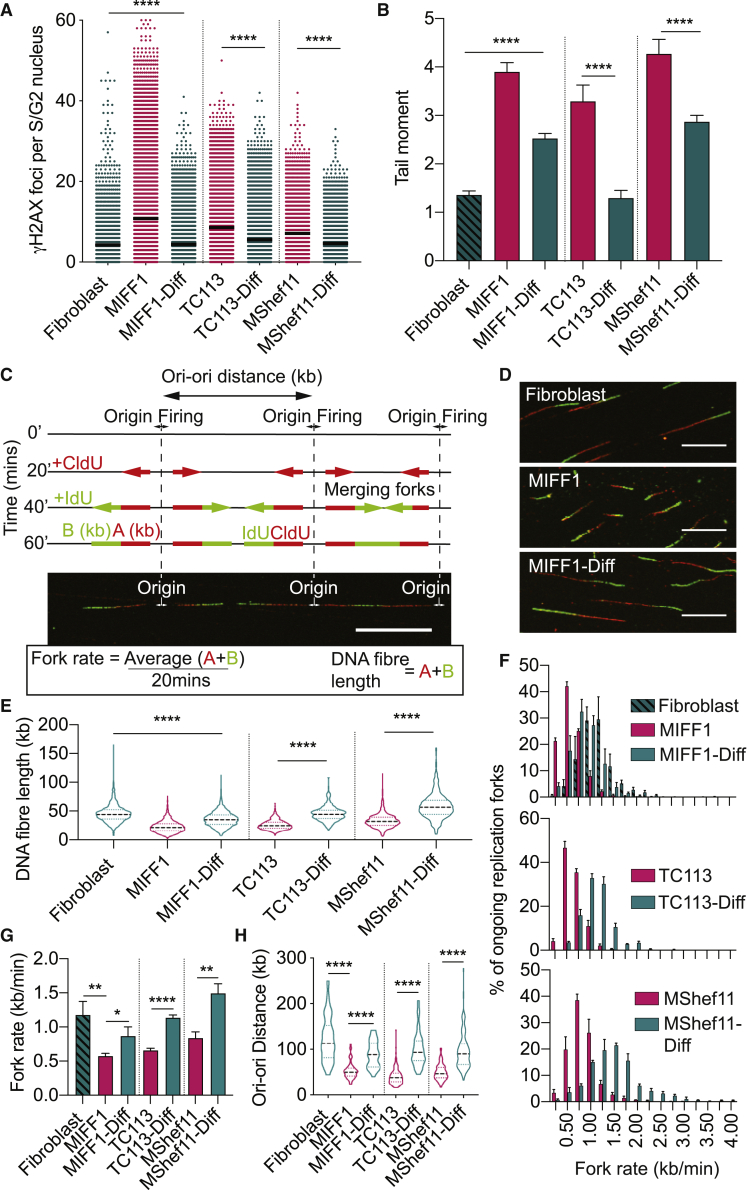
Figure 1.
Replication Stress and a Susceptibility to DNA Damage is a Characteristic of Undifferentiated Human PSCs
Data in the figure compare undifferentiated and differentiated cell states. MIFF1 was compared with its parent fibroblast line (Fibroblast) and with its differentiated derivative (MIFF1-Diff). TC113 and MShef11 were compared with their differentiated derivatives (TC113-Diff and MShef11-Diff).
(A) The number of γH2AX foci per S-/G2-phase cell. The S/G2 phase was determined from nuclear DNA content. Data points in (A) represent individual cells and are the results from three independent experiments; the center line indicates the mean. Two-tailed t test, ∗∗∗∗p < 0.0001 (n > 100 cells per cell line per experiment).
(B) Average tail moment from neutral comet assays. Data displayed are from three independent experiments ± SEM. Two-tailed t test, ∗∗∗∗p < 0.0001 (n ≥ 300 cells per cell line per experiment).
(C) Schematic of DNA fiber analysis. Sequential 20-min pulses of CldU and IdU labeled the progressing replication forks. Measurements of CldU and IdU lengths enable the analysis of replication fork dynamics. Scale bar, 10 μm.
(D) Representative DNA fibers are shown for Fibroblast, MIFF1, and MIFF1-Diff. Scale bar, 10 μm.
(E) Combined length of CldU and IdU in individual fibers (n > 200 forks per cell line per experiment, n = 3 experiments). Median distance, 25th and 75th quartiles are presented. Two-tailed t test, ∗∗∗∗p < 0.0001.
(F) Distribution of replication fork rates (n > 200 forks per cell line per experiment, n = 3 experiments). Data are mean values from each experiment ± SEM.
(G) Mean fork rates from (F) ± SD. Two-tailed t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 (n = 3 experiments).
(H) Distribution of adjacent origins distance measurements (Ori-ori). Median distance, 25th and 75th quartiles are presented. Two-tailed t test, ∗∗∗∗p < 0.0001 (n > 30 per cell line, n = 3 experiments).