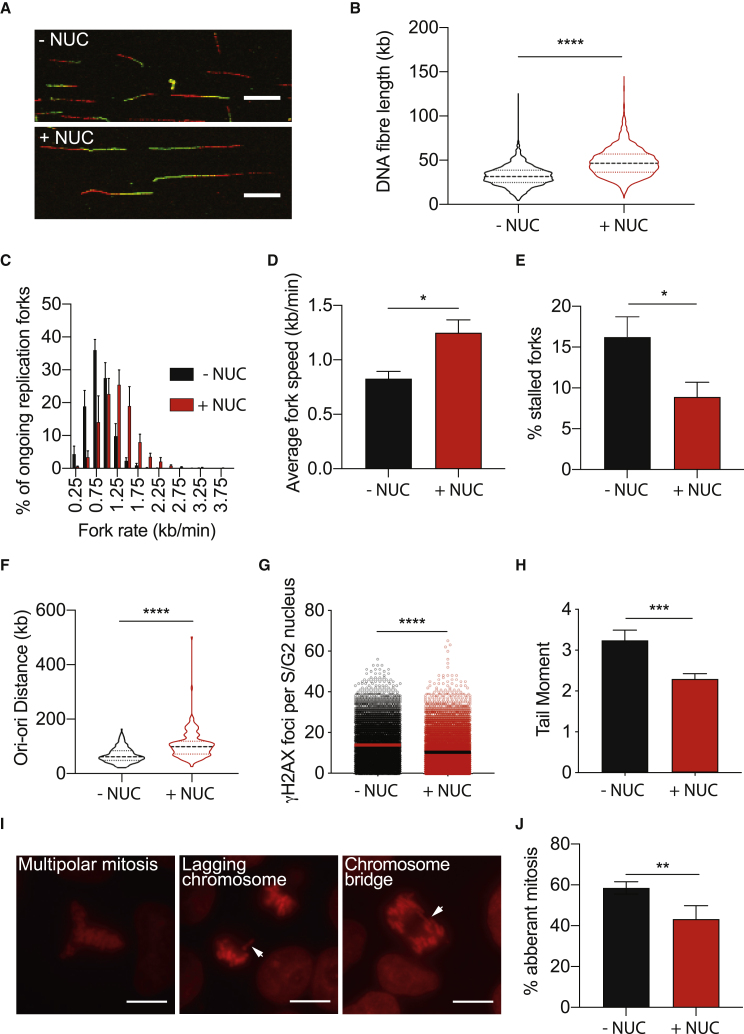
Figure 3.
Exogenously Supplied Nucleosides Alleviate Replication Stress in Human PSCs
(A) Representative images of DNA fibers in the absence of nucleosides (−NUC) and in the presence of exogenous nucleoside (+NUC) conditions. Scale bar, 10 μm.
(B) Combined length of CldU and IdU in individual fibers (n > 200 forks per cell line per experiment, n = 3 experiments). Median distance, 25th and 75th quartiles are presented. Two-tailed t test, ∗∗∗p < 0.001.
(C) Distribution of replication fork rates (n > 200 forks per condition per experiment, n = 3 experiments). Data are means ± SEM.
(D) Average fork rates from (C). Data are means ± SD. Two-tailed t test, ∗p < 0.05 (n = 3 experiments).
(E) Frequency of CldU-only tracts that denote a stalled replication fork (n > 700 forks per condition per experiment, n = 3 experiments). Data are means ± SD. Two-tailed t test, ∗p < 0.05.
(F) Distribution of adjacent origins distance measurements (Ori-ori). Median distance, 25th and 75th quartiles are presented. Two-tailed t test, ∗∗∗∗p < 0.0001 (n > 150 per cell line, n = 3 experiments).
(G) γH2AX foci per S-/G2-phase cell (determined from DNA content). Each data point is the measurement of an individual cell; the center line indicates the mean. Two-tailed t test, ∗∗∗∗p < 0.0001 (n > 100 cells per condition per experiment, n = 3 experiments).
(H) Average tail moment from neutral comet assay experiments. Data are means ± SD. Two-tailed t test, ∗∗∗p < 0.001 (n ≥ 300 cells per condition per experiment, n = 3 experiments).
(I and J) Mitotic errors observed from fluorescently labeled chromatin (histone H2B-RFP). Representative images of mitotic errors (I). White arrows point to mitotic error in each case. Scale bar, 10 μm. Average frequency of mitotic errors observed (J). Data are means ± SD. Unpaired t test, ∗∗p < 0.01 (n = 13–43 mitosis assessed per condition per experiment, n = 4 experiments).