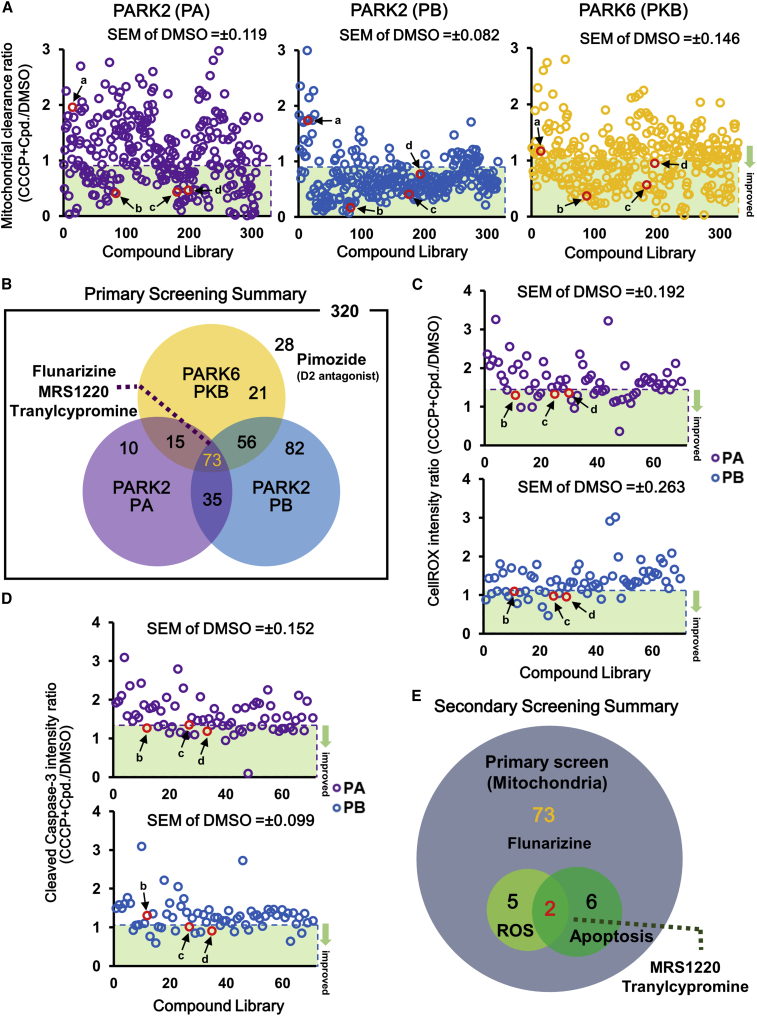
Figure 2.
Screening for Compounds that Modify Mitochondrial Clearance in PARK2 and PARK6 Neurons
(A) A scatterplot of mitochondrial clearance screening for PARK2 and PARK6 neurons. Data represent the ratio of mitochondrial area in neurons treated with CCCP and compound (Cpd) and that in neurons treated with DMSO. Hit compounds (below the average value of CCCP + DMSO treatment minus SEM) are indicated by the green band. Four notable compounds are indicated by the red frame. The average value ± SEM of CCCP + DMSO treatment: PA 1.06 ± 0.119, PB 1.00 ± 0.082, PKB 1.19 ± 0.146.
(B) Summary of the mitochondrial clearance screening (primary screening).
(C) A scatterplot of ROS accumulation screening for PARK2 neurons. Data represent the ratio of fluorescence intensity of CellROX in neurons treated with CCCP and compound and that in neurons treated with DMSO. Hit compounds (below the average value of CCCP + DMSO treatment minus SEM) are indicated by the green band. Three notable compounds are indicated by the red frame. The average value ± SEM of CCCP + DMSO treatment: PA 1.65 ± 0.192, PB 1.42 ± 0.263.
(D) A scatterplot of apoptosis screening for PARK2 neurons. Data represent the ratio of fluorescence intensity of cleaved caspase-3 treated with CCCP and compound to that treated with DMSO. Hit compounds (below the average value of CCCP + DMSO treatment minus SEM) are indicated by the green band. Three notable compounds are indicated by the red frame. The average value ± SEM of CCCP + DMSO treatment: PA 1.52 ± 0.152, PB 1.16 ± 0.099.
(E) Summary of ROS accumulation and apoptosis screening for PARK2 neurons (secondary screening).
CCCP, carbonyl cyanide 3-chlorophenylhydrazone; a, pimozide; b, flunarizine; c, MRS1220; d, tranylcypromine. See also Figure S3.