Abstract
Motivation
Genetic variation in regulatory elements can alter transcription factor (TF) binding by mutating a TF binding motif, which in turn may affect the activity of the regulatory elements. However, it is unclear which motifs are prone to impact transcriptional regulation if mutated. Current motif analysis tools either prioritize TFs based on motif enrichment without linking to a function or are limited in their applications due to the assumption of linearity between motifs and their functional effects.
Results
We present MAGGIE (Motif Alteration Genome-wide to Globally Investigate Elements), a novel method for identifying motifs mediating TF binding and function. By leveraging measurements from diverse genotypes, MAGGIE uses a statistical approach to link mutations of a motif to changes of an epigenomic feature without assuming a linear relationship. We benchmark MAGGIE across various applications using both simulated and biological datasets and demonstrate its improvement in sensitivity and specificity compared with the state-of-the-art motif analysis approaches. We use MAGGIE to gain novel insights into the divergent functions of distinct NF-κB factors in pro-inflammatory macrophages, revealing the association of p65–p50 co-binding with transcriptional activation and the association of p50 binding lacking p65 with transcriptional repression.
Availability and implementation
The Python package for MAGGIE is freely available at https://github.com/zeyang-shen/maggie. The accession number for the NF-κB ChIP-seq data generated for this study is Gene Expression Omnibus: GSE144070.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Genome-wide association studies (GWASs) have identified thousands of genetic variants associated with an increase in disease risk (MacArthur et al., 2017). Many of these variants fall within regulatory elements such as promoters and enhancers, implicating an effect on transcriptional regulation (Farh et al., 2015; GTEx Consortium et al., 2015; Khurana et al., 2016). Transcription factors (TFs) play an essential role in mediating the activity of regulatory elements. Many TFs possess DNA-binding domains that recognize specific DNA sequences, called TF binding motifs. Alterations of TF binding motifs have been established as an important mechanism for genetic variants to affect transcriptional regulation (Deplancke et al., 2016; Grossman et al., 2017; Heinz et al., 2013). However, it is not always straightforward which TF binding motifs are prone to have an impact on transcriptional regulation if mutated. First of all, a genetic variant is able to alter multiple motifs. Binding motifs for hundreds of TFs are currently available in the public databases (Fornes et al., 2020; Kulakovskiy et al., 2018; Matys et al., 2006; Weirauch et al., 2014). Many motifs correspond to similar or overlapping DNA sequences, which can be altered by the same variant simultaneously. The second complication is due to the strong dependency of TF binding on conditions. Multiple TF binding motifs are usually packed at regulatory elements across 100–200 base pairs (Lambert et al., 2018) but can become functional under different conditions depending on cell type, developmental time point, stimulus etc. (Spitz and Furlong, 2012) Knowing the function of motifs for a given condition can help prioritize TFs prone to be affected by genetic variation and ultimately have an impact on transcriptional regulation.
Numerous motif analysis tools have been published in the past decade to prioritize important TFs for experimental validation (Boeva, 2016; Jayaram et al., 2016). One major category of tools identifies enriched motifs that appear more frequently at given regions of interest than random genomic regions (Heinz et al., 2010; Machanick and Bailey, 2011; Siebert and Söding, 2016). Due to the development of high-throughput sequencing assays, these approaches can now be applied to various types of epigenomic features, such as chromatin accessibility measured by the assay for transposase-accessible chromatin using sequencing (ATAC-seq) or DNase I hypersensitive sites sequencing (DNase-seq), and TF binding and histone modification measured by chromatin immunoprecipitation sequencing (ChIP-seq) etc. (Reuter et al., 2015). However, this category of methods does not connect motif enrichment to a function, so the identified motifs may not have any functional impact on the epigenomic feature of interest.
Another category of motif analysis tools prioritize TFs by leveraging measurements and genetic variation of multiple human individuals or animal strains. Many of these methods depend on an assumption of linearity between the motif and the signal of epigenomic features (Fonseca et al., 2019; Mcvicker, 2013; Grubert et al., 2015; Link et al., 2018b). This assumption worked for TF binding but likely does not hold for many other epigenomic features like histone modification or stimulus response of regulatory elements, which result from the interactions between multiple TFs and may not possess a simple linear relationship with TF binding motifs.
Here, we developed a novel approach, MAGGIE (Motif Alteration Genome-wide to Globally Investigate Elements), to identify DNA motifs mediating TF binding and function. Considering the increasing amount of genotype and epigenomic data for different individuals and animal strains, we are able to identify genomic regions associated with a biased epigenomic feature of interest between different genotypes, labeling them as positive or negative for sequences with or without the feature, respectively (Fig. 1A). We propose to associate these biased regions with changes of TF binding motifs caused by genetic variation to gain insights into the functions of motifs. Unlike conventional motif enrichment methods, MAGGIE is independent of the background frequency of motifs and gains power in capturing the functional impacts of motifs by leveraging motif mutations at the same regions between individuals or strains. MAGGIE differs from other methods that associate motif mutations with epigenomic features by eliminating the assumption of linearity between motifs and testing features. The design of this framework is flexible in accommodating any type of epigenomic feature, including but not limited to the ones to be discussed in this article, such as TF binding, open chromatin, histone modification and stimulus response of regulatory elements.
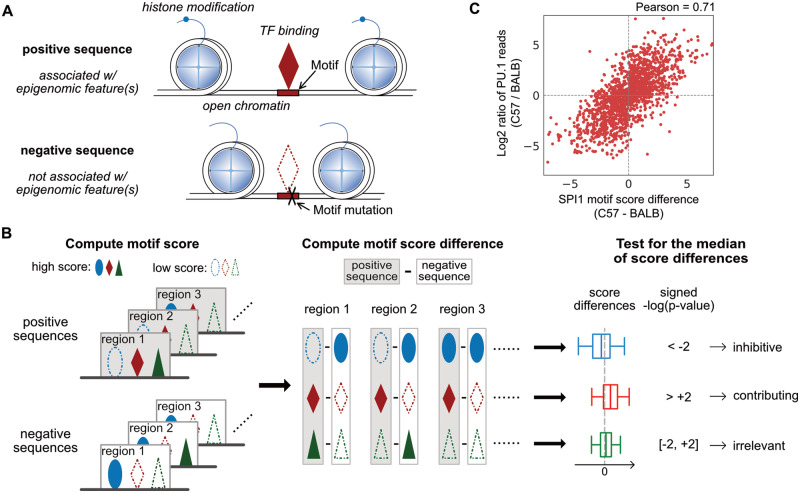
Fig. 1.
Overview of MAGGIE. (A) Schematic depicting how the epigenomic features of regulatory elements are related to the inputs of MAGGIE. Positive sequences are defined to be associated with epigenomic feature(s) of interest, such as TF binding, open chromatin, histone modification etc. Each positive sequence has a negative counterpart, which has a loss of the chosen epigenomic feature(s) due to mutations on TF binding motifs. (B) Flowchart of MAGGIE. Positive and negative sequences are used to compute motif scores as an estimated likelihood of being bound by certain TF. A representative motif score is obtained for each sequence by taking the maximum, displayed by different shapes (ellipse, diamond and triangle) for different TFs. High motif scores are shown as solid shapes and low scores as dashed shapes. Next, differences of representative motif scores are computed for every TF by subtracting scores of negative from positive sequences. Finally, the score differences for each TF are aggregated, and the median value is tested by Wilcoxon signed-rank test to evaluate whether there is a bias in the changing direction from positive to negative sequences. The examples demonstrate a significant bias of increase (ellipse) or decrease (diamond) or an insignificant bias (triangle), which implicates the inhibitive, contributing, or irrelevant role of TF, respectively. (C) Correlation between motif score differences of SPI1 motif and log2-fold changes of PU.1 binding activity between BALB and C57 mice. Each dot represents one of the 1641 PU.1 binding sites that have SPI1 motif mutations between the two strains
We evaluated the performance of MAGGIE in both simulated datasets and biological datasets and compared our results to HOMER (Heinz et al., 2010), MMARGE (Link et al., 2018b) and TBA (Fonseca et al., 2019), which are representative for the existing motif analysis tools. The results demonstrated the superior sensitivity and specificity of MAGGIE for detecting the effects of motif mutations in all of the experiments. By applying MAGGIE to the regulatory elements of macrophages in response to pro-inflammatory stimulus, we captured divergent functions of distinct NF-κB (nuclear factor-kappa B) factors despite the similarity of their motifs. These results were further validated by the NF-κB binding sites measured by ChIP-seq experiments, showing the promise of MAGGIE in identifying highly specific motifs and discovering novel functions of TFs.
2 Materials and methods
2.1 Overview of MAGGIE
The overall framework of MAGGIE is illustrated in Figure 1B. MAGGIE takes pairs of sequences as inputs. Positive sequences are identified to be associated with an epigenomic feature of interest, while negative sequences are from different alleles or the same regions of a different genome where the epigenomic feature is not found. Depending on the genetic difference of genomes, every pair of input sequences can have a variable number of genetic variants like single-nucleotide polymorphisms (SNPs) and short insertions and deletions.
The basic assumption for MAGGIE is that the allele specificity of an epigenomic feature is derived from the genetic variation between positive and negative sequences that mutate TF binding motifs. This assumption is supported by the findings that motif mutations due to local genetic variation is the major explanation for the gain or loss of TF binding sites (Kundaje et al., 2015; Link et al., 2018a). Considering the importance of TFs for other epigenomic features like promoter and enhancer function (Reiter et al., 2017; Spitz and Furlong, 2012), we hypothesized that our framework could help identify motifs mediating both TF binding and other epigenomic features affected by TF binding.
The computation of MAGGIE is centered on the motif score based on position weight matrix (PWM), which is the widely used metric to approximate the likelihoods of being bound by certain TF (Stormo, 2000). Given pairs of positive and negative sequences associated with a chosen allele-specific epigenomic feature, MAGGIE computes motif scores for hundreds of TFs whose PWMs are currently available in the JASPAR database (Fornes et al., 2020). For each TF, a representative motif score is calculated for every sequence by taking the maximal score across the sequence. MAGGIE then computes differences of representative motif scores by subtracting scores of negative from positive sequences to obtain the changes of binding likelihood. Score differences should have a bias toward positive values (i.e. higher motif scores in positive sequences) if the corresponding TF is contributing to the chosen epigenomic feature. On the contrary, if the TF is potentially inhibitive for the chosen feature, the aggregated differences will tend to have negative values (i.e. lower motif scores in positive sequences). Irrelevant TFs will have their motifs randomly mutated by genetic variation, so the score differences should be overall balanced around zero. A non-parametric Wilcoxon signed-rank two-sided test is used to statistically test the significance of the association between motif mutations and the chosen epigenomic feature by asking whether the median of all the non-zero motif score differences is close to zero. A signed P-value combining the sign of median score difference with the P-value from statistical tests implicates the function of TF to be either contributing (positive) or inhibitive (negative) if called significant.
2.2 Computation of motif score and motif score difference
Motif score is a reliable metric to measure the likelihood of TF binding and can well reflect the binding activity of the corresponding TF (Boeva, 2016; Ji et al., 2018). A PWM stores the log likelihoods for the four possible nucleotides (A, C, G and T) to be bound by a TF at each position (Stormo, 2000):
where is the probability of seeing nucleotide n at the kth position of the motif, and is the background probability for different nucleotides. Given a DNA sequence, we can compute motif scores for any TF by adding up the log likelihoods of seeing certain nucleotides at every position:
where is the motif score for a segment of the given sequence from position i to position i + L−1, supposing L is the length of the motif and i starts at 1, and is the nucleotide at position i + k. For a sequence longer than the motif (i.e. the biggest possible i > L), instead of dealing with a list of motif scores, we obtain the maximal motif score to represent the binding likelihood of the entire sequence:
where r is the starting position of the maximal motif score. Every sequence pair will yield two representative motif scores whose starting positions are notated by and for positive and negative sequence, respectively:
Then, the log-fold change of binding likelihood within the sequence pair can be computed by subtracting the representative motif score of the negative sequence from that of the positive sequence:
If we set the background probability as the same for the four types of nucleotides (i.e. 0.25), the difference of representative motif score turns out to be the log-fold change of the binding likelihood between positive and negative sequences:
Here, we compute the motif score difference based on the maximal score of each sequence, which may or may not at the same location ( not necessarily equal to ). This strategy is able to compensate for the effects from nearby variants and the interactions between multiple motifs. Any representative motif score less than zero is replaced by zero before computing a score difference in order to reduce impacts from poorly matched motifs. Motif score difference has been used as an indicator of the change in TF binding (Martin et al., 2019; Spivakov et al., 2012). For example, by comparing PU.1 binding in macrophages of C57BL/6J (C57) and BALB/cJ (BALB) mice (Link et al., 2018a), we observed a strong positive correlation between the score difference of SPI1 motif and the change in PU.1 (encoded by SPI1) binding quantified by ChIP-seq reads (Fig. 1C). This relationship is independent of the actual motif score (Supplementary Fig. S1). We saw a diminished correlation using non-uniform background probabilities (Supplementary Fig. S2) or restricting motifs at the same locations () instead of their respective best matches (Supplementary Fig. S3). These intrinsic characteristics of motif score difference support the hypotheses that (i) motif score difference can indicate change in binding of the corresponding TF, and (ii) aggregated motif score differences can reflect whether the presence of specific epigenomic feature is associated with the gain or loss of TF binding.
2.3 Applications and data preparation
2.3.1. Simulated data
To characterize the performance of MAGGIE and systematically compare with other methods, we conducted simulated experiments. Positive sequences were generated by first randomly selecting A, C, G or T to form sequences of 200-base pair (bp). Then we created TF binding motifs by sampling nucleotides based on their probabilities derived from PWMs and inserted these motifs at non-overlapping random positions. To obtain counterpart negative sequences, SNPs were simulated inside hypothetic ‘contributing’ motifs by changing the existing nucleotides.
During the generation of simulated data, we inserted ‘irrelevant’ motifs, which experienced either no mutation or random mutation, to evaluate the specificity of MAGGIE. The sensitivity of MAGGIE was tested by changing the number of simulated sequences (i.e. sample size) or the fraction of sequences having motif mutations [i.e. signal-to-noise ratio (SNR)].
2.3.2. TF binding sites
We tested MAGGIE to identify TF binding motifs for corresponding TF binding. Allele-specific binding sites of 12 TFs were obtained from two cell types, GM12878 and HeLa-S3 (Shi et al., 2016). We extracted 100-bp sequences around the SNPs associated with allele-specific binding sites and labeled the sequences with the binding alleles as positive sequences and those with the non-binding alleles as negative.
MAGGIE was then used to identify collaborative TFs. We downloaded the ChIP-seq data of PU.1 and C/EBPβ for C57 and BALB mice from the Gene Expression Omnibus (GEO) database with accession number GSE109965 (Link et al., 2018a), and the ChIP-seq data of ATF3 for the same mouse strains with the accession number GSE46494 (Fonseca et al., 2019). The data for C57 were mapped to the mm10 genome using Bowtie2 v2.3.5.1 (Langmead and Salzberg, 2012), whereas the data for BALB were first mapped to the BALB genome and then shifted to the mm10 genome using the MMARGE v1.0 ‘shift’ function (Link et al., 2018b). The reproducible TF binding sites were identified by using HOMER v4.9.1 to call unfiltered 200-bp peaks (Heinz et al., 2010) and running IDR v2.0.3 on replicates with the default parameters (Li et al., 2011). The TF binding sites found only in one of the strains were defined to be strain-specific, yielding 13099 PU.1, 8127 C/EBPβ and 13347 ATF3 strain-specific binding sites between BALB and C57. The sequences of strain-specific binding sites were extracted from both strains using the MMARGE v1.0 ‘extract_sequences’ function (Link et al., 2018b). Sequences associated with TF binding are labeled as positive regardless of which strain they are originated from, and their counterpart sequences from the other strain are labeled as negative.
2.3.3. Chromatin quantitative trait loci
We applied MAGGIE to discover motifs mediating chromatin accessibility and histone modification. DNaseI sensitivity quantitative trait loci (dsQTLs) were downloaded from the GEO database with accession number GSE31388 (Degner et al., 2012). Histone QTLs (hQTLs) were acquired for three types of histone modifications, local acetylation of histone H3 lysine 27 (H3K27ac), mono-methylation of histone H3 lysine 4 (H3K4me1) and tri-methylation of histone H3 lysine 4 (H3K4me3; Grubert et al., 2015). All QTLs were originally analyzed for lymphoblastoid cell lines (LCLs). We obtained more stringent hQTLs based on a P-value < 1e-6 and a distance from the associated peak <1000 bp. QTLs were further separated based on HOMER annotations into promoter, intronic and distal subsets to investigate functional motifs of different genomic regions. Distal QTLs are those within intergenic regions and >2000 bp from the nearest transcription start sites. Similar to the pre-processing for the allele-specific binding sites, we extracted 100-bp sequences centering around the variants and labeled the alleles associated with a higher trait level as positive and the other alleles as negative.
2.3.4. Stimulus responses of regulatory elements
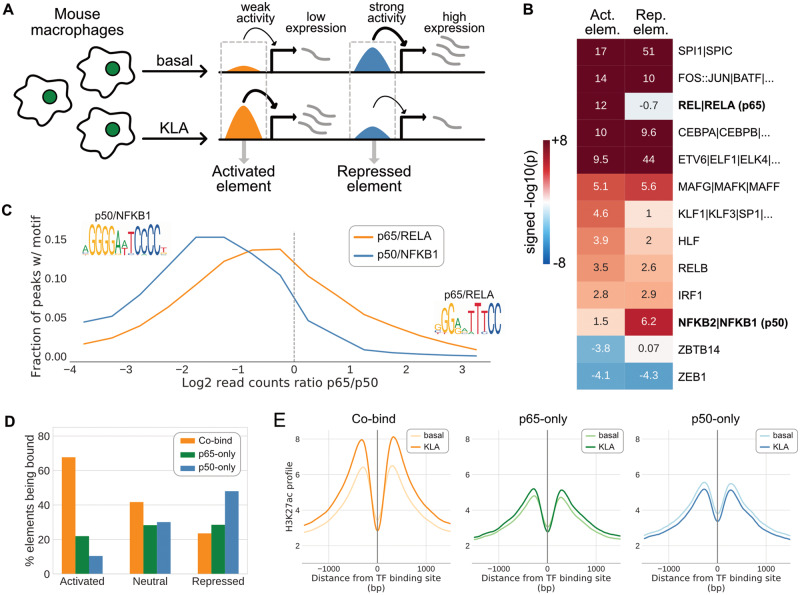
The application of MAGGIE was further extended to the stimulus response of regulatory elements. We downloaded ATAC-seq and H3K27ac ChIP-seq data from macrophages at both basal state and pro-inflammatory state induced by 1-h treatment of the TLR4-specific ligand Kdo2 lipid A (KLA) from four diverse strains of mice: C57BL/6J (C57), NOD/ShiLtJ (NOD), PWK/PhJ (PWK) and SPRET/EiJ (SPRET; Link et al., 2018a). Similar to the pre-processing of ChIP-seq data for TFs, the raw reads were mapped and shifted to the mm10 genome. Based on ATAC-seq data, we obtained 200-bp reproducible open chromatin and filtered for intergenic and intronic regions to obtain potential enhancers. Open chromatin regions of the two conditions from the same strain were merged and extended from 200 to 1000 bp to quantify their activity by the count of H3K27ac ChIP-seq reads. We filtered for active regulatory elements (>16 reads in at least one condition; Supplementary Fig. S6) and computed the change of activity from basal to KLA-treated condition by the fold change of reads. Regions showing a higher or lower level of H3K27ac >2.5-fold after KLA treatment were labeled as ‘activated’ or ‘repressed’, respectively (Fig. 4A), and those with <40% change were labeled as ‘neutral’. Based on pairwise comparisons across the four mouse strains, regulatory elements labeled as ‘activated’ or ‘repressed’ only in one of the compared strains were called strain-specific and were pooled for analysis.
Fig. 4.
Divergent functions of NF-κB factors in pro-inflammatory macrophages captured by MAGGIE and validated by experiments. (A) Sketch of KLA-activated and KLA-repressed regulatory elements defined by >2.5-fold changes of H3K27ac from basal to KLA-treated conditions. (B) Significant functional motifs identified by MAGGIE for activated and repressed regulatory elements. Similar motifs are separated by ‘|’ and shown with their average results. Protein names of RELA and NFKB1 are shown in the brackets, corresponding to p65 and p50, respectively. (C) Binding activities of NF-κB factors associated with RELA or NFKB1 motifs. Motifs were searched within the 200-bp binding sites of p65 and p50 at the KLA-treated condition measured by ChIP-seq experiments. ChIP-seq reads for both p65 and p50 were counted to quantify binding activities. Regions with at least 32 reads of either factor were used to compute the log2 ratio of reads between p65 and p50. The distributions of log ratios are displayed in orange for sites having RELA motif (10 549 sites) and in blue for sites having NFKB1 motif (2144 sites). The logos of motif PWMs are demonstrated. (D) Co-existence of NF-κB binding and the KLA responses of regulatory elements. NF-κB binding sites were separated into sites bound by p65 alone (p65-only), p50 alone (p50-only) or both (Co-bind). Among the regulatory elements that overlap with NF-κB binding sites, the bar plots summarized the fractions of elements bound by different NF-κB factors for activated, neutral, and repressed elements. (E) Change of H3K27ac at NF-κB binding sites after KLA treatment. H3K27ac ChIP-seq reads were counted within ±1500 bp around the three categories of NF-κB binding sites using a bin size of 25 bp and were averaged to show the overall change of H3K27ac profiles
2.4 Comparative methods
We compared the performance of MAGGIE against several existing methods in identifying functional TF binding motifs. The most obvious competitors are those that also leverage measurements from diverse genotypes, including a recently developed method called MMARGE (Link et al., 2018b), which fits a linear mixed model between the motif score and the signal of epigenomic features (e.g. TF binding activity). Unlike other approaches based on linear assumptions, MMARGE additionally corrects for individual variance while leveraging genetic variation between individuals. MMARGE v1.0 was downloaded from https://github.com/vlink/marge. Since the existing linear methods do not directly work with binary-labeled datasets (e.g. simulated data, QTLs), we implemented a replacement model that fit motif scores against binary labels in the simulated experiments using statsmodels package (Seabold and Perktold, 2010).
Another big category of motif analysis tools is based on motif enrichment algorithms, such as HOMER (Heinz et al., 2010), MEME Suite (Machanick and Bailey, 2011), BaMM (Siebert and Söding, 2016) etc. We performed comparisons between enriched and functional motifs identified by MAGGIE. We expect any one of those methods to be representative for the others, so we picked HOMER in our experiments, which was downloaded from http://homer.ucsd.edu/homer/data/software/homer.v4.9.1.zip. Besides using HOMER to find enriched motifs, we extended its application to calculate differential enrichment between positive and negative sequences and evaluated the performance of enrichment algorithms in detecting motif mutations.
We also adapted a machine learning-based approach, TBA, to detect motif mutations between positive and negative sequences (Fonseca et al., 2019). We trained a logistic regression model with representative motif scores, from which P-values were generated from the likelihood-ratio test to represent the importance of each motif in classifying binary labels. The model training modules were downloaded from https://github.com/jenhantao/tba. All of the comparative methods above were run with the default parameters. Since none of these methods output signed P-values as MAGGIE does, we reported only P-values from MAGGIE in any comparative studies.
2.5 Validation experiment
Bone marrow was isolated from C57 mice and differentiated for 7 days using media containing M-CSF to generate bone marrow-derived macrophages (BMDMs) as described previously (Link et al., 2018a). BMDMs were maintained at basal conditions or treated with KLA for 1 h. For p65 (Santa Cruz, sc-372X) and p50 (Abcam, ab32360) ChIP-seq experiments, 8 million untreated or KLA-treated BMDMs per assay were double-crosslinked using disuccinimidyl glutarate and formaldehyde (FA). ChIP-seq was performed using 2 μg of antibody as described previously (Heinz et al., 2018). ChIP DNA was prepared for sequencing using the NEBNext Ultra II DNA library prep kit (NEB, E7645) and sequencing was performed on the HiSeq4000 (75 bp SR, Illumina). The binding sites of p65 and p50 were identified using HOMER ‘findPeaks -size 200’ (Heinz et al., 2010) and then merged to obtain co-binding sites and p65- or p50-only binding sites. The binding activity of p65 and p50 was quantified by the count of ChIP-seq reads. The raw and processed data have been deposited to the NCBI GEO under the accession number GSE144070.
3 Results
3.1 MAGGIE shows superior specificity and sensitivity on simulated datasets
To evaluate the performance of MAGGIE, we stochastically simulated one thousand DNA sequences of 200 bp embedded with an arbitrary pair of motifs, SPI1 and CEBPB, labeled as positive sequences. Negative sequences were then derived from this set by switching single nucleotides of the SPI1 motif for half of the positive sequences. Table 1 shows the top motifs output from MAGGIE and three comparative approaches: linear model, logistic regression adapted from TBA and HOMER. Both MAGGIE and linear model identified SPI1 and its similar motifs as the most significant hits. Logistic regression that was trained to classify positive and negative sequences lacked the sensitivity to detect SPI1 motif. On the contrary, HOMER identified both SPI1 and CEBPB as significantly enriched over the default random backgrounds for both positive (‘pos versus bg’ column) and negative sequences (‘neg versus bg’ column). As expected, enriched motifs failed to distinguish the mutated motif from the unmutated motif, which was only captured by methods that leveraged motif mutations resulted from synthetic genetic variation.
Table 1.
Top motifs output from different motif analysis tools evaluated on the simulated datasets
| Rank | MAGGIE | Linear model | Logistic regression (TBA) | HOMER—pos versus bg | HOMER—neg versus bg |
|---|---|---|---|---|---|
| 1 | SPI1* (13) | SPI1* (8.6) | SPI1 (1.0) | CEBPG* (198) | CEBPG* (195) |
| 2 | SPIB* (10) | SPIB* (6.5) | ZSCAN10 (0.8) | CEBPD* (194) | CEBPB* (183) |
| 3 | SPIC* (4.7) | ETV6 (3.6) | EWSR1-FLI1 (0.8) | CEBPB * (192) | CEBPD* (181) |
| 4 | ETV6* (4.5) | SPIC (3.6) | STAT5A (0.6) | CEBPE* (191) | CEBPE* (178) |
| 5 | ELF1* (4.2) | EHF (3.2) | SIX6 (0.4) | SPI1* (177) | SPI1* (127) |
Note: −log10 P-values are shown in parentheses. (*) indicates motifs that passed FDR < 0.05 after the Benjamini–Hochberg controlling procedure. As the true positive, SPI1 motif is highlighted in bold.
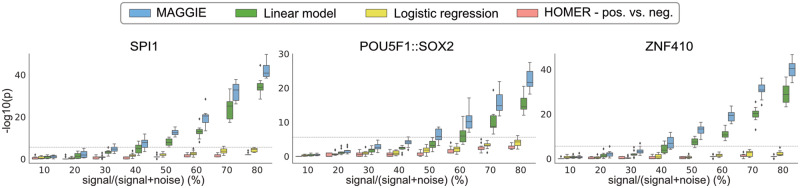
To assess the sensitivity of MAGGIE, we tested its performance when different fractions of sequences experienced motif mutations (i.e. SNR). For every SNR ranging from 10% to 80%, we repeated simulation of sequences 10 times and aggregated P-values for embedded motifs from the comparative methods. Here, we also assessed the performance of the motif enrichment algorithm implemented by HOMER in detecting motif mutations by setting positive sequences as inputs and negative sequences as backgrounds. We evaluated the comparative methods on three arbitrary pairs of motifs: SPI1-CEBPB, POU5F1::SOX2-KLF4 and ZNF410-IRF1. For each experiment, one motif pair was inserted into sequences, but only the first motif (SPI1, POU5F1::SOX2 and ZNF410) was mutated by synthetic genetic variation. MAGGIE consistently outperforms the other methods in identifying the mutated motif (Fig. 2) and not the unmutated motif (Supplementary Fig. S4). Even though the other methods could potentially pass the significance threshold with a higher SNR or an increasing sample size (Supplementary Fig. S5), MAGGIE showed superior sensitivity when motifs are mutated in <50% of the finite samples.
Fig. 2.
Comparison of sensitivity between MAGGIE and other approaches on simulated datasets. Each boxplot aggregates the significance values from 10 simulations. Boundary lines show the median and quartiles of each distribution. Every simulation generated a thousand sequences inserted with a pair of motifs, which serve as the positive set. 10–80% of these sequences had a single nucleotide changed within the SPI1 motif for SPI1-CEBPB pair, or the POU5F1::SOX2 motif for POU5F1::SOX2-KLF4 pair or the ZNF410 motif for ZNF410-IRF1 pair, whereas the rest were kept untouched, resulting in the negative set. The dashed lines indicate the significance threshold after multiple testing correction
3.2 MAGGIE identifies known mediators for TF binding sites and QTLs
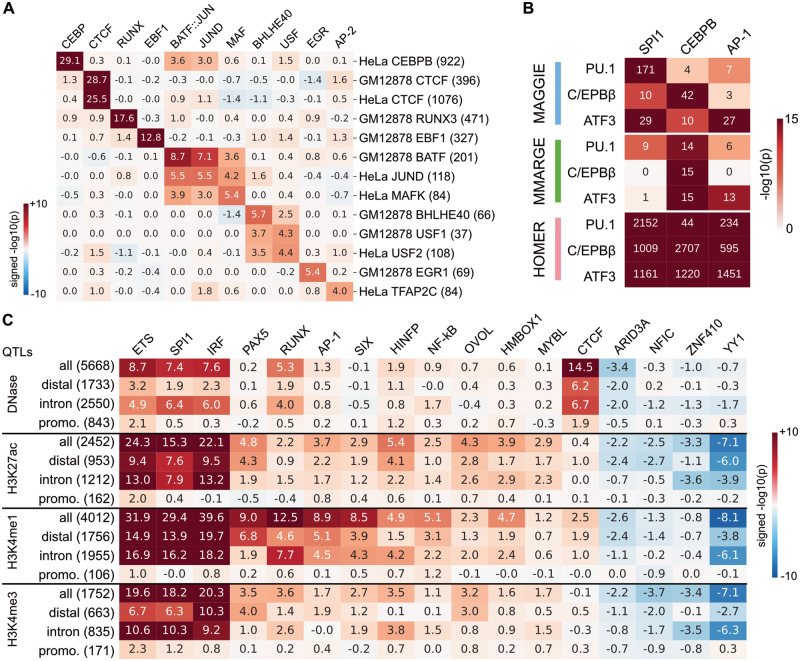
After observing the superior performance of MAGGIE on simulated data, we tested our method with several biological datasets. First, we analyzed the allele-specific TF binding sites associated with SNPs (Shi et al., 2016). Among the 13 experiments tested, MAGGIE identified the corresponding motifs of the bound TFs for all of them (Fig. 3A). Even though P-values vary due to the quality and the sample size of each dataset, the corresponding motifs were recognized as the most significant even for TFs with as few as 37 allele-specific binding sites like USF1.
Fig. 3.
Functional motifs identified by MAGGIE for various epigenomic features using biological datasets. (A) Signed P-values for allele-specific TF binding sites. In total, 13 datasets were analyzed covering 12 different TFs from two cell types: GM12878 and HeLa-S3 (HeLa). Datasets are arranged vertically with their sample sizes displayed in brackets, and motifs are shown horizontally on top by their gene names. (B) Comparative results for strain-specific TF binding sites. P-values from different motif analysis methods are shown for the corresponding motifs of the three LDTFs (PU.1, C/EBPβ and ATF3). (C) Significant motifs identified for chromatin QTLs of LCLs. Signed P-values from MAGGIE are shown for the entire sets as well as the subsets based on the locations of QTLs. The number of QTLs in each set is shown in brackets. Motifs shown here were tested significant for at least one type of the QTLs. All the results in this figure have been averaged for similar motifs and are displayed by their family names (e.g. ETS, AP-1)
Next, we evaluated whether MAGGIE is able to recover the collaborative binding between TFs. Regulatory elements are usually bound by multiple TFs together, which form a complex with other co-activators to regulate functions (Reiter et al., 2017). For example, lineage-determining TFs (LDTFs) of macrophages such as PU.1, C/EBP, and AP-1 factors were frequently found to co-bind at macrophage-specific enhancers (Glass and Natoli, 2016; Heinz et al., 2015). Previous studies showed that the binding of specific LDTFs was not only dependent on each factor’s own motif, but also on nearby motifs recognized by collaborative factors (Heinz et al., 2013; Link et al., 2018a). To verify this conclusion with MAGGIE, we downloaded ChIP-seq data for PU.1 (encoded by SPI1), C/EBPβ (encoded by CEBPB) and ATF3, which binds to AP-1 motif, from two genetically diverse strains of mice, C57BL/6J (C57) and BALB/cJ (BALB; Fonseca et al., 2019; Link et al., 2018a). Strain-specific TF binding sites were identified for each factor and analyzed with MAGGIE. As comparison, we used MMARGE to find motifs correlated with TF binding activity quantified by ChIP-seq read counts and used HOMER to find enriched motifs among positive sequences in comparison to random backgrounds. The outputs from the three approaches for the relevant motifs are summarized in Figure 3B. MAGGIE recognized PU.1 binding to be mostly dependent on its own motif instead of any other motif, while C/EBPβ binding was highly dependent on CEBPB motif but also significantly dependent on SPI1 motif. The results were consistent with the pioneer role of PU.1 in opening chromatin and guiding the binding of other TFs (Barozzi et al., 2014). On the contrary, the comparative methods failed to distinguish the different functions between the bound TF and its collaborative factors. HOMER assigned strong significance to all the motifs because it was designed to identify enriched motifs without considering functions. MMARGE showed a lack of power in detecting collaborative factors using the data of two mouse strains as it requires more data or larger genetic difference to confidently identify a correlation between motif and TF binding.
The general framework of MAGGIE can also be applied to QTL datasets for epigenomic features that are influenced by TF binding. We downloaded QTLs of several epigenomic features for LCLs, including dsQTLs for chromatin accessibility (Degner et al., 2012) and hQTLs for three types of histone modifications, H3K27ac, H3K4me1 and H3K4me3 (Grubert et al., 2015). MAGGIE identified motifs with different specificity for the testing features (Fig. 3C). CTCF was output at top for dsQTLs but was insignificant for each type of hQTLs, supporting the major role of CTCF in maintaining chromatin structures instead of inducing active chromatin states (Arzate-Mejía et al., 2018). PU.1 together with other ETS factors were significant for both chromatin accessibility and histone modifications, indicating a pioneer role in opening chromatin as well as an important role in activating regulatory elements in LCLs (Scott et al., 1994). MAGGIE also identified many other motifs important for histone modifications, which have been found to maintain the cell identity and function of LCLs from previous studies, such as PAX5 (Glimcher and Singh, 1999), RUNX (Mevel et al., 2019) and NF-κB (Nagel et al., 2014). It is intriguing that several motifs showed up with potentially inhibitive functions, although these will need to be confirmed in later studies.
3.3 MAGGIE captures divergent functions of NF-κB factors for the stimulus responses of regulatory elements
Next, we tested MAGGIE with a more complex epigenomic feature: stimulus responses of regulatory elements. ATAC-seq and H3K27ac ChIP-seq data from four genetically diverse strains of mice were downloaded for macrophages at basal conditions and at pro-inflammatory conditions induced by 1-h treatment of KLA (Link et al., 2018a). We used ATAC-seq data to locate open chromatin regions accessible for TF binding and H3K27ac ChIP-seq data to quantify the activity of these regions and identify active regulatory elements (Supplementary Fig. S6). By filtering for 2.5-fold change of activity from basal to KLA-treated conditions, we identified KLA-activated and KLA-repressed regulatory elements for each mouse strain (Fig. 4A). Among those, ∼12 000 activated elements and 18 000 repressed elements were specific to one of the strains based on pairwise comparisons. Strain-specific activated and repressed regulatory elements were separately tested by MAGGIE to identify their mediators. Interestingly, besides SPI1, CEBP and AP-1 (e.g. FOS::JUN) motifs that were known to be important for the KLA responses of macrophages (Glass and Natoli, 2016), MAGGIE assigned divergent functions for NF-κB factors (Fig. 4B). RELA corresponding to p65 subunit was output as functional for the activation response, while NFKB1 corresponding to p50 subunit was found significant for the repressed elements. On the contrary, due to the similarity of these motifs (Supplementary Fig. S7), HOMER found both motifs enriched in the activated elements compared with random backgrounds and neither enriched in the repressed elements (Supplementary Fig. S8). Previous studies have shown that p65 frequently forms heterodimers with p50 to act as a transcriptional activator, while p50 homodimers result in transcriptional repression (Brignall et al., 2019; Cheng et al., 2011; Natoli et al., 2005). However, the genome-wide functions and binding patterns of these factors remain unknown.
To validate the functions of p65 and p50 for the KLA responses of macrophages, we conducted ChIP-seq experiments in C57 mice for p65 and p50 to measure their genome-wide binding sites in KLA-treated macrophages. Based on the measured TF binding sites, we first investigated the binding patterns of these factors. We searched for sites with RELA or NFKB1 motifs and computed the binding activities of NF-κB factors at those sites by counting ChIP-seq reads. Regions with relatively strong binding of either factor (>32 ChIP-seq reads of p65 or p50; Supplementary Fig. S9) were used to calculate the log ratios of read counts between p65 and p50 (Fig. 4C). RELA motif was enriched at the co-binding sites of p65 and p50, while NFKB1 motif was more strongly bound by p50. To connect the binding patterns to the regulatory elements used in MAGGIE, we overlapped the TF binding sites with the activated and repressed elements previously defined for C57 mice and found that the majority of activated elements were co-bound by both p65 and p50, while repressed elements were more often bound by p50 alone (Fig. 4D). By quantifying the regulatory activity around the binding sites of p65 and p50 by the level of H3K27ac, we found an overall decrease in H3K27ac around sites only bound by p50 and an overall increase around the co-binding sites of p65 and p50 after KLA treatment (Fig. 4E). These findings suggest a genome-wide role of p65–p50 heterodimers as a transcriptional activator and p50 homodimers as a repressor for KLA-treated macrophages. More importantly, our experimental results validated the predictions from MAGGIE regarding the divergent functions of p65 and p50 subunits for pro-inflammatory macrophages, showing promise of using MAGGIE to discover novel functions of TFs for complex epigenomic features.
4 Discussion
To our knowledge, MAGGIE is the first work to associate the mutation of TF binding motif with various types of epigenomic features. In contrast to motif enrichment methods, such as HOMER, in which identified motifs may or may not be functionally related to epigenomic features, MAGGIE determines the significance of motifs based on putative functional consequences of local motif mutations. Due to this qualitative difference, MAGGIE and motif enrichment methods recover overlapping but non-identical sets of significant motifs. As the major difference in methodology, MAGGIE focuses on the change of motif score and intentionally ignores the actual motif score due to the strong correlation between motif mutation and change of TF binding (Fig. 1C) and the independency of this relationship from the actual motif score (Supplementary Fig. S1). Another reason not to incorporate the actual motif score is that many epigenomic features do not possess a simple relationship with motif score. Instead of assuming a linear relationship between motif scores and epigenomic features like many current methods do, MAGGIE tests for a bias in the changing direction of motif mutations. We demonstrated that MAGGIE is able to identify known functional motifs for TF binding (Fig. 3A and B), chromatin accessibility (Fig. 3C), and histone modification (Fig. 3C). MAGGIE also helped to discover divergent functions of distinct NF-κB factors for the KLA response of regulatory elements in macrophages (Fig. 4), which was not found by any other motif analysis tools. It is worth noting that the motifs of NF-κB factors are usually too similar to be distinguished by motif enrichment methods, but the strategy of focusing on the change of motif score instead of the actual motif score is sensitive enough to capture the difference.
MAGGIE takes binary-labeled datasets as inputs (i.e. positive and negative sequences), which facilitates application to most publicly available data, including aggregated datasets like QTLs and processed data from sequencing experiments like ChIP-seq and ATAC-seq. However, for the framework to work, MAGGIE requires additional measurements and genetic variation information for at least two different genotypes, which may not be currently available for some biological problems. The primary limitation to the discovery power of MAGGIE is the degree of genetic variation provided by the samples being analyzed. Another limitation is the inevitable cutoff accompanied with binary labels, which might affect the results especially when there are concerns about insufficient sample size or low data quality.
The flexibility of our statistical framework makes it applicable to any type of epigenomic feature that is potentially affected by TF binding. Given the stand-alone tool we provided for the motif analysis methods described here, it will be interesting to investigate the performance of MAGGIE in other features, such as chromatin interaction and DNA methylation. Another future extension is to switch the PWM score used in this study to other types of motif scores, such as more advanced representations of TF binding motifs based on hidden Markov models. It will also be promising to incorporate state-of-the-art machine learning approaches (e.g. deep learning) into our framework to consider complex interactions between motifs. For instance, we can integrate the prediction scores of variant impacts from deep learning models and associate those predictions with biased changes of motifs.
In summary, we presented a novel method for identifying DNA sequence motifs mediating TF binding and function, which goes beyond enriched or correlated motifs that are frequently analyzed nowadays. Given the growing interest in the function of TFs and the unprecedented generation of epigenomic data for different individuals and animal strains, we expect MAGGIE to be an effective bioinformatic tool that can be included in the regular routine of motif analysis.
Supplementary Material
Acknowledgements
We would like to express our great appreciation to Melissa Gymrek, Jenhan Tao and Ludmil B. Alexandrov for friendly reviews. Our special thanks are extended to Inge R. Holtman for beta testing and feedback on the software package, and Jana Collier for technical assistance.
Funding
This work was supported by the following grants: National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases R01 DK091183 and Foundation Leducq Grant 16CVD01. M.A.H. was also supported by a Rubicon grant from the NWO (Netherlands Organization for Scientific Research) and a postdoctoral grant from the Amsterdam Cardiovascular Sciences (ACS) institute.
Conflict of Interest: none declared.
References
- Arzate-Mejía R.G. et al. (2018) Developing in 3D: the role of CTCF in cell differentiation. Development, 145, dev137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I. et al. (2014) Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell, 54, 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V. (2016) Analysis of genomic sequence motifs for deciphering transcription factor binding and transcriptional regulation in eukaryotic cells. Front. Genet., 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignall R. et al. (2019) Considering abundance, affinity, and binding site availability in the NK-κB target selection puzzle. Front. Immunol., 10, 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.S. et al. (2011) The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci. Signal., 4, ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J.F. et al. (2012) DNase I sensitivity QTLs are a major determinant of human expression variation. Nature, 482, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B. et al. (2016) The genetics of transcription factor DNA binding variation. Cell, 166, 538–554. [DOI] [PubMed] [Google Scholar]
- Farh K.K.-H. et al. (2015) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature, 518, 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca G.J. et al. (2019) Diverse motif ensembles specify non-redundant DNA binding activities of AP-1 family members in macrophages. Nat. Commun., 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O. et al. (2020) JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res., 48, D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K., Natoli G. (2016) Molecular control of activation and priming in macrophages. Nat. Immunol., 17, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher L.H., Singh H. (1999) Transcription factors in lymphocyte development—T and B cells get together. Cell, 96, 13–23. [DOI] [PubMed] [Google Scholar]
- Grossman S.R. et al. (2017) Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc. Natl. Acad. Sci. USA, 114, E1291–E1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubert F. et al. (2015) Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell, 162, 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium et al. (2015) The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. (2013) Effect of natural genetic variation on enhancer selection and function. Nature, 503, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. (2015) The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol., 16, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. (2018) Transcription elongation can affect genome 3D structure. Cell, 174, 1522–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram N. et al. (2016) Evaluating tools for transcription factor binding site prediction. BMC Bioinformatics, 17, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z. et al. (2018) Genome-scale identification of transcription factors that mediate an inflammatory network during breast cellular transformation. Nat. Commun., 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E. et al. (2016) Role of non-coding sequence variants in cancer. Nat. Rev. Genet., 17, 93–108. [DOI] [PubMed] [Google Scholar]
- Kulakovskiy I.V. et al. (2018) HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res., 46, D252–D259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A. et al. ; Roadmap Epigenomics Consortium. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S.A. et al. (2018) The human transcription factors. Cell, 172, 650–665. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V.M. et al. (2018. a) Analysis of genetically diverse macrophages reveals local and domain-wide mechanisms that control transcription factor binding and function. Cell, 173, 1796–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V.M. et al. (2018. b) MMARGE: motif mutation analysis for regulatory genomic elements. Nucleic Acids Res., 46, 7006–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J. et al. (2017) The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res., 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics, 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V. et al. (2019) QBiC-Pred: quantitative predictions of transcription factor binding changes due to sequence variants. Nucleic Acids Res., 47, W127–W135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res., 34, D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G. et al. (2013) Identification of Genetic Variants That Affect Histone Modifications in Human Cells. Science, 342, 747–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel R. et al. (2019) RUNX transcription factors: orchestrators of development. Development, 146, dev148296. [DOI] [PubMed] [Google Scholar]
- Nagel D. et al. (2014) Mechanisms and consequences of constitutive NF-κB activation in B-cell lymphoid malignancies. Oncogene, 33, 5655–5665. [DOI] [PubMed] [Google Scholar]
- Natoli G. et al. (2005) Interactions of NF-κB with chromatin: the art of being at the right place at the right time. Nat. Immunol., 6, 439–445. [DOI] [PubMed] [Google Scholar]
- Li Q. et al. (2011) Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat., 5, 1752–1779. [Google Scholar]
- Reiter F. et al. (2017) Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev., 43, 73–81. [DOI] [PubMed] [Google Scholar]
- Reuter J.A. et al. (2015) High-throughput sequencing technologies. Mol. Cell, 58, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.W. et al. (1994) Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science, 265, 1573–1577. [DOI] [PubMed] [Google Scholar]
- Seabold S., Perktold J. (2010) Statsmodels: econometric and statistical modeling with python. In: Proceedings of the 9th Python in Science Conference, Vol. 57, p. 61. Scipy. [Google Scholar]
- Shi W. et al. (2016) Evaluating the impact of single nucleotide variants on transcription factor binding. Nucleic Acids Res., 44, 10106–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert M., Söding J. (2016) Bayesian Markov models consistently outperform PWMs at predicting motifs in nucleotide sequences. Nucleic Acids Res., 44, 6055–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Furlong E.E. (2012) Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet., 13, 613–626. [DOI] [PubMed] [Google Scholar]
- Spivakov M. et al. (2012) Analysis of variation at transcription factor binding sites in drosophila and humans. Genome Biol., 13, R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G.D. (2000) DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- Weirauch M.T. et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell, 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.