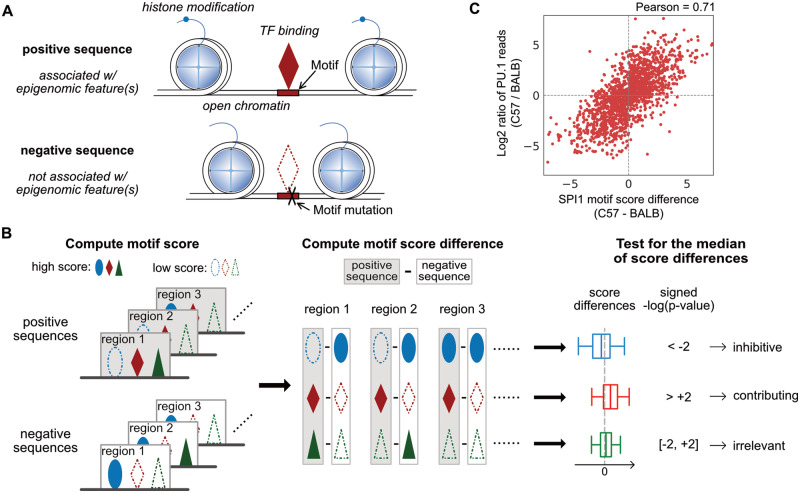
Fig. 1.
Overview of MAGGIE. (A) Schematic depicting how the epigenomic features of regulatory elements are related to the inputs of MAGGIE. Positive sequences are defined to be associated with epigenomic feature(s) of interest, such as TF binding, open chromatin, histone modification etc. Each positive sequence has a negative counterpart, which has a loss of the chosen epigenomic feature(s) due to mutations on TF binding motifs. (B) Flowchart of MAGGIE. Positive and negative sequences are used to compute motif scores as an estimated likelihood of being bound by certain TF. A representative motif score is obtained for each sequence by taking the maximum, displayed by different shapes (ellipse, diamond and triangle) for different TFs. High motif scores are shown as solid shapes and low scores as dashed shapes. Next, differences of representative motif scores are computed for every TF by subtracting scores of negative from positive sequences. Finally, the score differences for each TF are aggregated, and the median value is tested by Wilcoxon signed-rank test to evaluate whether there is a bias in the changing direction from positive to negative sequences. The examples demonstrate a significant bias of increase (ellipse) or decrease (diamond) or an insignificant bias (triangle), which implicates the inhibitive, contributing, or irrelevant role of TF, respectively. (C) Correlation between motif score differences of SPI1 motif and log2-fold changes of PU.1 binding activity between BALB and C57 mice. Each dot represents one of the 1641 PU.1 binding sites that have SPI1 motif mutations between the two strains