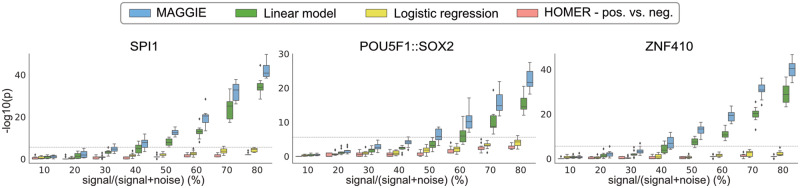
Fig. 2.
Comparison of sensitivity between MAGGIE and other approaches on simulated datasets. Each boxplot aggregates the significance values from 10 simulations. Boundary lines show the median and quartiles of each distribution. Every simulation generated a thousand sequences inserted with a pair of motifs, which serve as the positive set. 10–80% of these sequences had a single nucleotide changed within the SPI1 motif for SPI1-CEBPB pair, or the POU5F1::SOX2 motif for POU5F1::SOX2-KLF4 pair or the ZNF410 motif for ZNF410-IRF1 pair, whereas the rest were kept untouched, resulting in the negative set. The dashed lines indicate the significance threshold after multiple testing correction