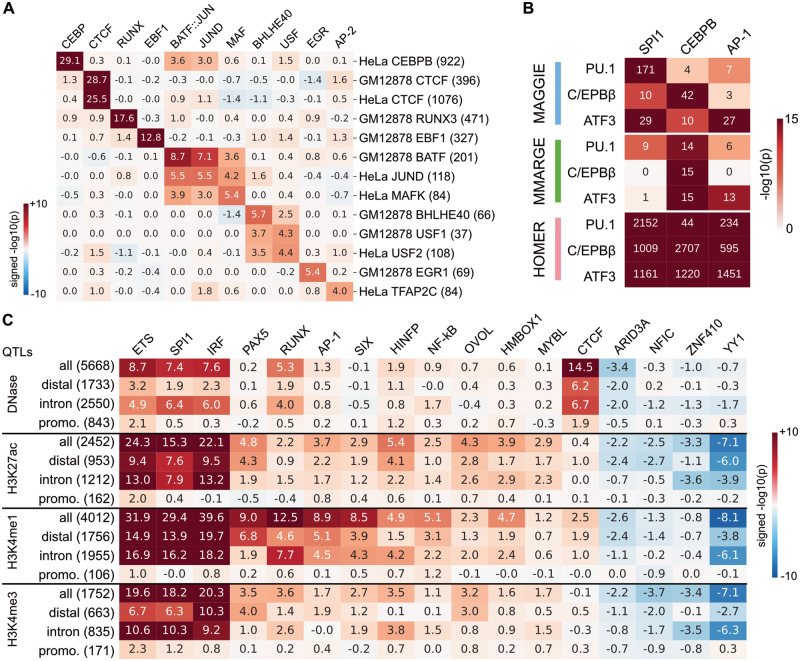
Fig. 3.
Functional motifs identified by MAGGIE for various epigenomic features using biological datasets. (A) Signed P-values for allele-specific TF binding sites. In total, 13 datasets were analyzed covering 12 different TFs from two cell types: GM12878 and HeLa-S3 (HeLa). Datasets are arranged vertically with their sample sizes displayed in brackets, and motifs are shown horizontally on top by their gene names. (B) Comparative results for strain-specific TF binding sites. P-values from different motif analysis methods are shown for the corresponding motifs of the three LDTFs (PU.1, C/EBPβ and ATF3). (C) Significant motifs identified for chromatin QTLs of LCLs. Signed P-values from MAGGIE are shown for the entire sets as well as the subsets based on the locations of QTLs. The number of QTLs in each set is shown in brackets. Motifs shown here were tested significant for at least one type of the QTLs. All the results in this figure have been averaged for similar motifs and are displayed by their family names (e.g. ETS, AP-1)