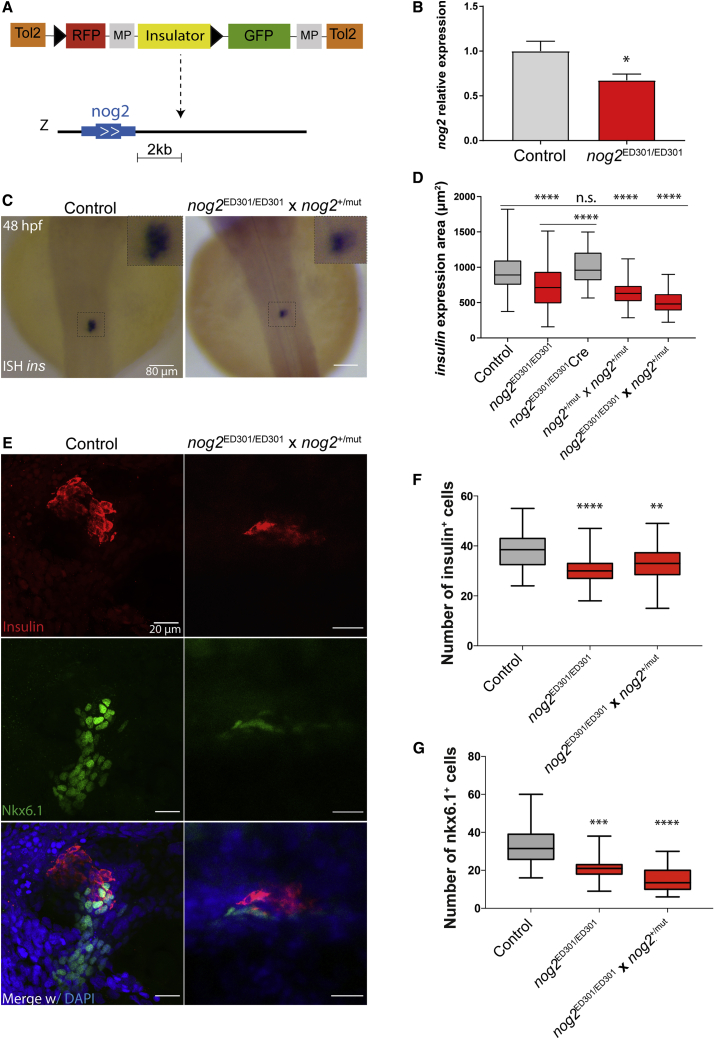
Figure 1.
nog2 Is Required for Proper Pancreas Development
(A) ED301 zebrafish line corresponds to an ED transposon integration containing a potent enhancer blocking insulator (yellow) and mapped 2 kb downstream of nog2. The loxP sequences are depicted as black triangles (see also Figure S1). Scale bar represents 2 kilobases.
(B) In the ED301 zebrafish line, nog2 expression is downregulated, as detected by qPCR performed in three biological replicates of 30 embryos each (error bars represent SD; ∗p < 0.05).
(C) In situ hybridization of insulin in 48 hpf representative embryos from an outcross of nog2ED301/ED301 and nog2+/mut lines compared with control embryos. Scale bar represents 80 μm.
(D) Quantification of the insulin expression area detected by in situ hybridization in control (n = 194), nog2 ED301/ED301 (n = 93), nog2ED301/ED301 injected with Cre recombinase (n = 32), nog2+/mut incross (n = 115), and nog2ED301/ED301, nog2+/mut outcross embryos (n = 36) at 48 hpf. Error bars represent SD; ∗∗∗∗p < 0.0001.
(E) Representative confocal images of 48 hpf zebrafish embryos counterstained with a DAPI nuclear marker (blue), an anti-insulin antibody marking β cells (red), and an anti-Nkx6.1 antibody marking pancreatic progenitor cells (green). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 20 μm.
(F) Quantification of the number of insulin-expressing cells in nog2ED301/ED301 and nog2ED301/ED301, nog2+/mut outcross embryos compared with controls (n ≥ 30). Error bars represent SD; ∗∗∗∗p < 0.0001, ∗∗p < 0.01.
(G) Quantification of the number of nkx6.1-expressing cells in nog2 ED301/ED301 and nog2ED301/ED301, nog2+/mut outcross embryos compared with controls (n ≥ 18). Error bars represent SD; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001.