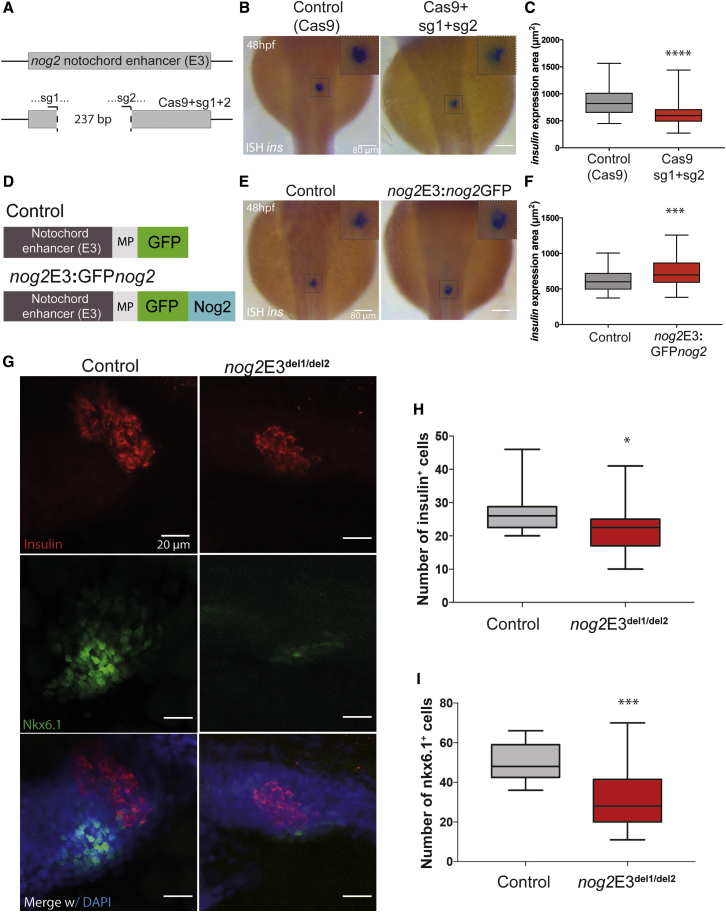
Figure 4.
The nog2 Notochord Enhancer Is Required for Proper Pancreas Development
(A) Representation of the WT nog2E3 enhancer (above) and somatic deletions (below) generated by the injection of the Cas9 protein, together with sg1 and sg2 targeting two regions of the sequence 237 bp apart (see also Figure S3).
(B) In situ hybridization of insulin in 48 hpf representative embryos, injected with the Cas9 protein, and two sgRNAs compared with control embryos, injected with only Cas9. Scale bars represent 80 μm.
(C) Quantification of the insulin expression area detected by in situ hybridization in injected embryos at 48 hpf compared with controls. Error bars represent SD; ∗∗∗∗p < 0.0001. In all cases, n ≥ 97.
(D) Diagram of the nog2E3:GFPnog2 construct used to achieve notochord-specific overexpression of GFPnog2 and the respective control.
(E) In situ hybridization of insulin in 48 hpf representative embryos injected with nog2E3:GFPnog2 compared with control embryos. Scale bars represent 80 μm.
(F) Quantification of the insulin expression area detected by in situ hybridization in injected embryos at 48 hpf compared with controls. Error bars represent SD; ∗∗∗p < 0.001. In all cases, n ≥ 38.
(G) Representative confocal images of 48 hpf zebrafish embryos counterstained with a DAPI nuclear marker (blue), an anti-insulin antibody marking β cells (red), and an anti-Nkx6.1 antibody marking pancreatic progenitor cells (green). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 20 μm.
(H) Quantification of the number of insulin-expressing cells in nog2E3del1/del2 embryos compared with controls (n ≥ 19). Error bars represent SD; ∗p < 0.05.
(I) Quantification of the number of nkx6.1-expressing cells in nog2E3del1/del2 embryos compared with controls (n ≥ 13). Error bars represent SD; ∗∗∗p < 0.001.