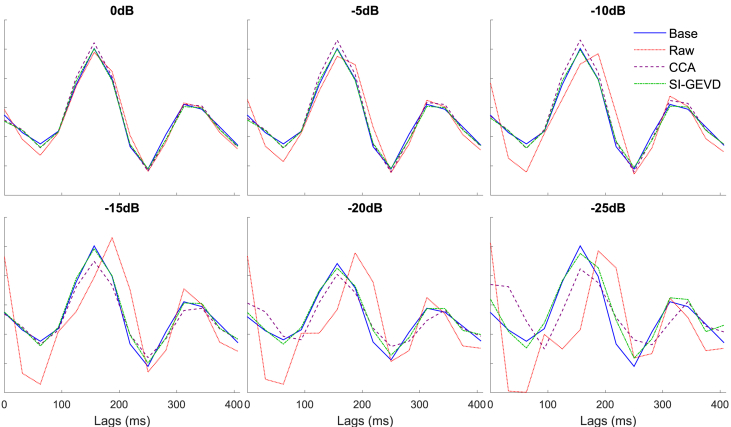
Results: For each 120 s trial, the estimated TRFs of all channels were concatenated into a single vector estimated TRF:
from the raw EEG data,
from the CCA-filtered data, and
from the SI-GEVD filtered EEG data. For each trial, the base TRFs of all channels (depending on which condition the trial belonged to, i.e., left or right attended) were also concatenated to obtain a single vector base TRF
. We omit the indication for the left or right attended stimulus conditions for notational convenience. In order to eliminate any differences due to scaling, a scaling factor was estimated by applying a least squares fitting such that the estimated TRF vector (referred to, in general, as
was scaled to fit
in the minimum mean squared error sense. This allows for a more fair comparison, as it allows to compensate for amplitude bias in the back-projection,
4 and since we are mainly interested in the shape and
relative amplitude of the TRFs across channels. The scaling was performed across channels (note that
w is a vector in which the TRFs of all channels are concatenated), and thus the relative differences in the per-channel amplitudes remained unchanged. This scaling factor was found as follows: