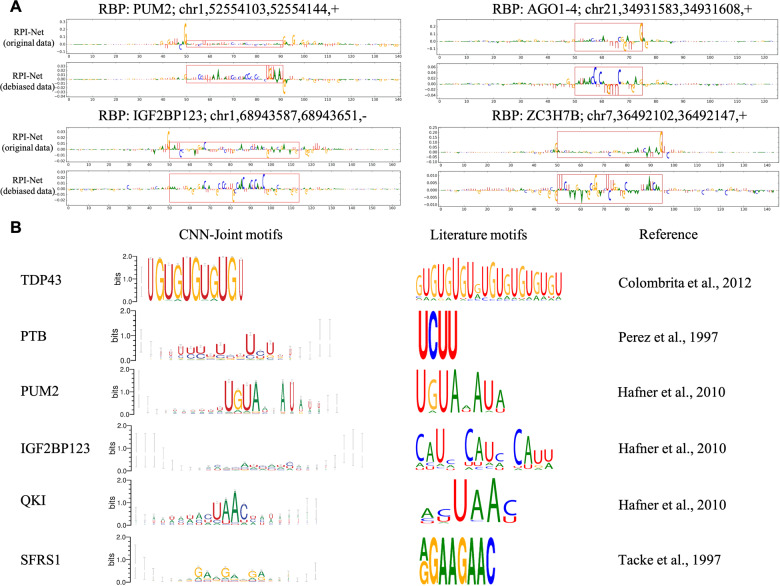
Fig. 4.
Integrated gradient maps and sequence motifs obtained using RPI-Net(CNN). (A) Integrated gradient values for four positive examples, revealing that debiasing the CLIP-Seq data is crucial for RPI-Net(CNN) to learn meaningful protein binding hypothesis. When RPI-Net is trained on the original (biased) datasets, the border artefacts will receive the highest attribution of the model’s prediction. Trained on debiased datasets, the border artefacts are eliminated and meaningful protein binding pattern are revealed inside the viewpoint region (enclosed red rectangles). (B) Shows six sequence logos extracted from RPI-Net(CNN) models, compared to the known motifs for those RBPs. The widths of characters become smaller as more gaps are inserted to that position. Our sequence motifs have shown a strong agreement to the literature motifs that are experimentally verified. Image credits: The literature motif images are taken from Maticzka et al. (2014), Figure 5