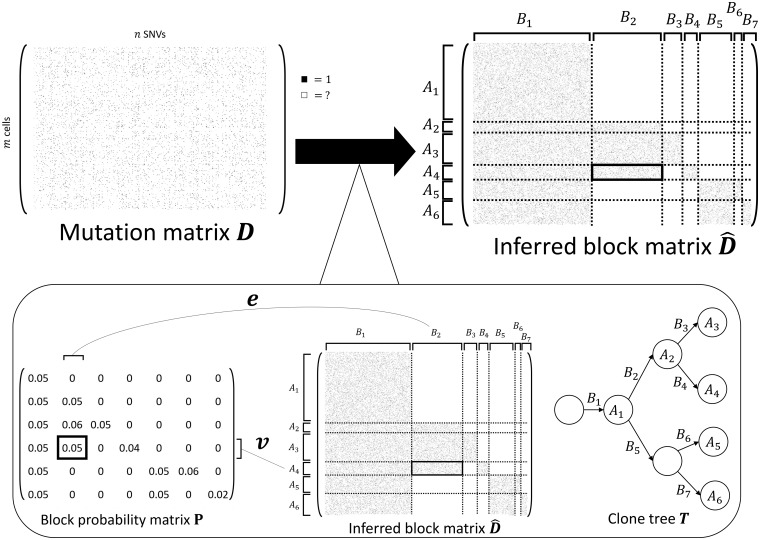
Fig. 1.
SBMClone solves the CBI problem using an SBM. (Top) An example mutation matrix D with m cells (rows) and n mutations (columns) is composed of entries with either 1 or ‘?’ to indicate the measurement (black square) or lack of measurement (white square) of a mutation in each cell. The goal of the CBI problem is to infer the rearrangement of the rows and columns of D that form the inferred block matrix where rows are partitioned into k row blocks (k = 6 here), columns are partitioned into column blocks ( here), and each block of has either high (many black squares) or low (no black squares) proportions of 1s. (Bottom left) SBMClone uses an SBM which is a generative model parameterized by the block probability matrix . Each block probability indicates the expected proportion of 1s within the block composed of the cells in row block Ar and the mutations in column block Bs. (Bottom right) The mutation matrix D has a block structure because every row block Ar corresponds to a clone, which accumulate clusters of mutations that correspond to every column block Bs; this evolutionary process is described as the clone tree T where every node correspond to a clone and every edge is labeled by a mutation cluster