Fig. 1.
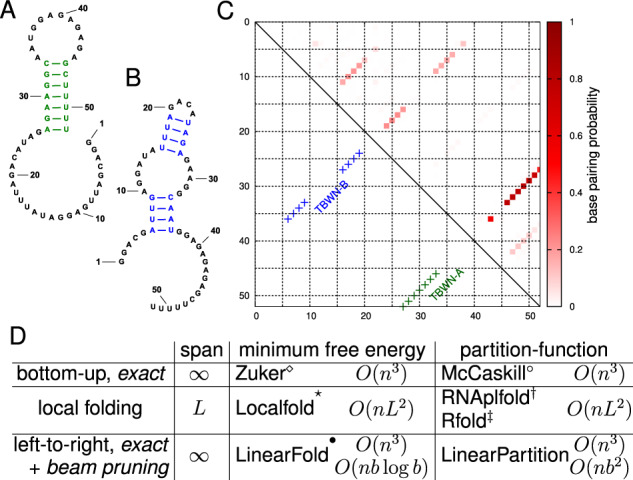
An RNA can fold into multiple structures at equilibrium. (A and B) Two secondary structures of Tebowned RNA: TBWN-A and TBWN-B (Cordero and Das, 2015). (C) Upper triangle shows the estimated base-pairing probability matrix for this RNA using Vienna RNAfold, where darker red squares represent higher probability base pairs; the lower triangle shows the two different structures; (D) Comparison between classical, local, and left-to-right algorithms for MFE and partition function calculation. (Zuker and Stiegler, 1981), °(McCaskill, 1990), *(Lange et al., 2012), (Bernhart et al., 2006a), (Kiryu et al., 2008) and (Huang et al., 2019). LinearFold and LinearPartition enjoy linear runtime because of a left-to-right order that enables heuristic beam pruning, and both become exact algorithms without pruning. ‘Span’ denotes the window size (max. pair distance) ( means no limit); it is a small constant in local methods (e.g. default L= 70 nt in RNAplfold)
