Fig. 4.
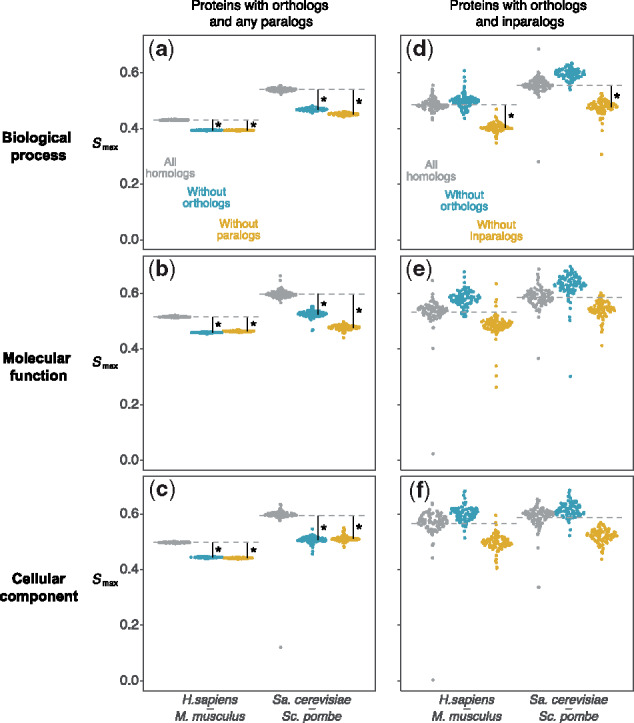
The impact of excluding orthologs or paralogs from prediction. values for proteins having orthologs and one other homolog (panels a-c) and proteins having orthologs and inparalogs (panels d-f), in human–mouse (Hs–Mm) and Sa.cerevisiae–Sc.pombe (Sc–Sp), are lower when ignoring either homolog. Points are 100 bootstrap samples of each protein set, and asterisks indicate a significant decrease in (bootstrap P < 0.05)
