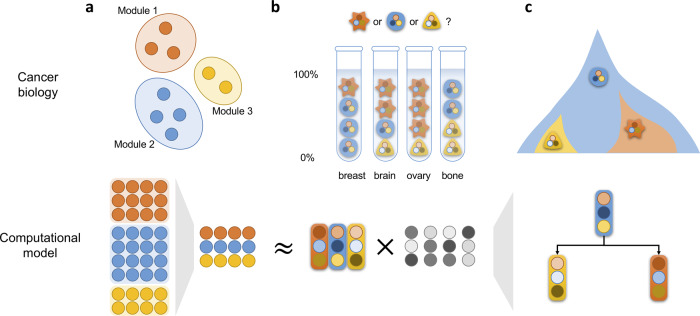
Fig. 1.
Illustration of discovering underlying cancer evolutionary mechanisms of metastatic progression using the proposed computational models. (a) Gene module compression. Genes from the same modules have similar coexpression patterns. By mapping the individual genes into modules, we can get a cleaner representation of bulk RNA data. (b) Deconvolution of bulk RNA. Using the RAD algorithm, we unmixed the bulk data into two matrices: expression matrix and fraction matrix, which represent the expression profile and fractions of individual cell populations. (c) Phylogeny inference of cell communities. The inferred phylogeny represents the most likely evolutionary trajectories of cell populations