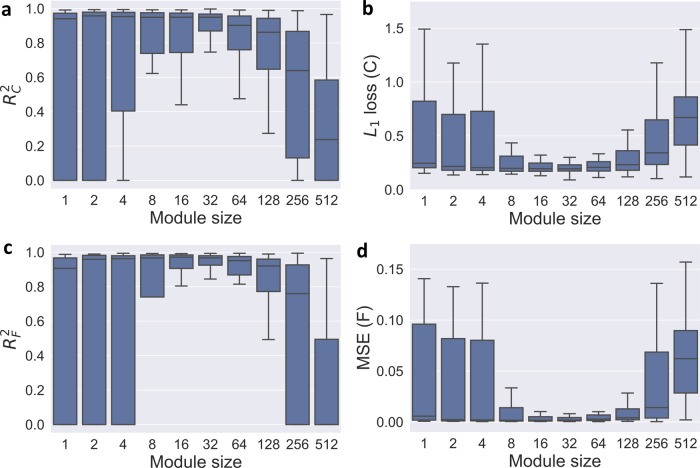
Fig. 2.
Effectiveness of gene module representation. Compressing the expression of individual genes into gene modules can promote more robust deconvolution with proper module size. We tested on the simulated bulk data with the noise level of σ = 4, and repeated all the experiments for 100 times to get the boxplot. We evaluated four metrics that measure the accuracy of deconvolution with different gene module sizes. Note that ‘module size’ of one is equivalent to the original gene expression without module compression. (a) Accuracy of expression matrix estimation: . (b) Error of expression matrix estimation: L1 loss. (c) Accuracy of fraction matrix estimation: . (d) Error of fraction matrix estimation: MSE