Fig. 5.
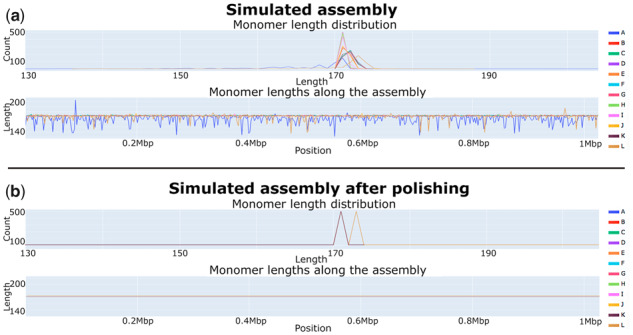
Monomer length distribution for the simulated (a) and simulatedpolish (b) assemblies. Monomer sequences forming a consensus DXZ1* sequence, derived in Bzikadze and Pevzner (2019), were used for analysis. In the simulated assembly, the length of the A-monomers varies from 131 to 203 bp (mean 165 bp) and the length of the L-monomers varies from 137 to 187 bp (mean 171 bp). In the simulatedpolish assembly, the length of all A-monomers (L-monomers) is equal to 171 (173) bp. Since all monomers, except for L, have lengths 171 bp after polishing, they all are represented by the color corresponding to the K-monomer
