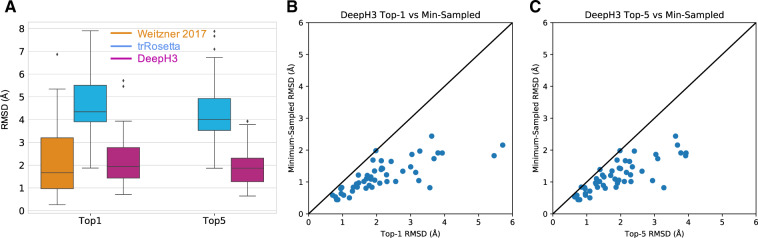
Fig. 6.
Performance of DeepH3 for de novo CDR H3 loop structure prediction. (A) DeepH3 achieves lower average RMSD (2.2 ± 1.1 Å) than trRosetta (4.7 ± 1.4 Å) and ties Weitzner et al. (2.2 ± 1.5 Å) (Weitzner et al., 2017) when the best scoring structures for each target were compared (top 1). When the lowest-RMSD structure among the five best-scoring structures were considered (top 5), DeepH3 (1.9 ± 0.9 Å) outperformed trRosetta (4.3 ± 1.3 Å). Top-5 metrics were not available for Weitzner et al. (B) Comparison of the minimum RMSD sampled by DeepH3 to the RMSD of the best-scoring structure (top 1) for each target. (C) Comparison of the minimum RMSD sampled by DeepH3 to the lowest RMSD within the set of five best-scoring structures (top 5) for each target.