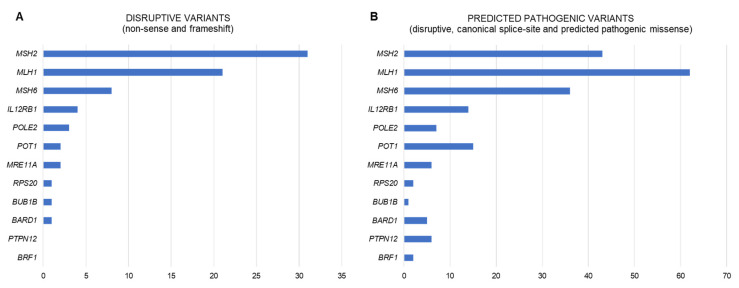
Figure 2.
(A) Number of disruptive (non-sense and frameshift) and (B) predicted pathogenic (disruptive, canonical splice-site, and predicted pathogenic missense) variants identified in the MMR genes MLH1, MSH2 and MSH6, RPS20, and in the most promising candidate genes for nonpolyposis CRC predisposition, in 1006 familial/early onset CRC cases. PMS2 was not included in the graph due to the possibility of miscalled variants due to the existence of multiple pseudogenes. Data obtained from Chubb et al., 2016 [28].